Search Count: 85
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: MLT, SO4 |
 |
The Crystal Structure Of Full-Length Pak2 Containing K278R And D368N Mutants
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of Human Glutaminyl Cyclase In Complex With N-(2-(1H-Imidazol-5-Yl)Ethyl)-4-Methoxybenzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: TRANSFERASE Ligands: ZN, A1EFY |
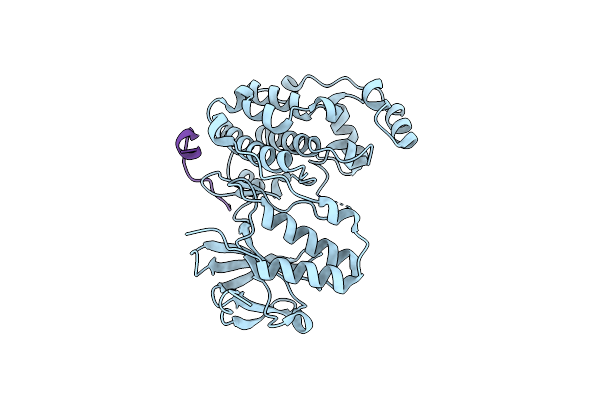 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-03-19 Classification: TRANSFERASE |
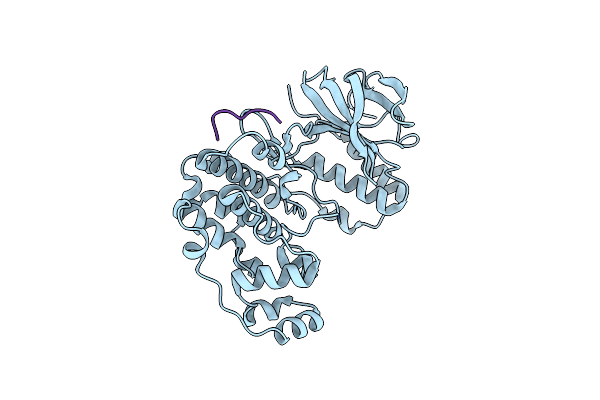 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-03-19 Classification: TRANSFERASE |
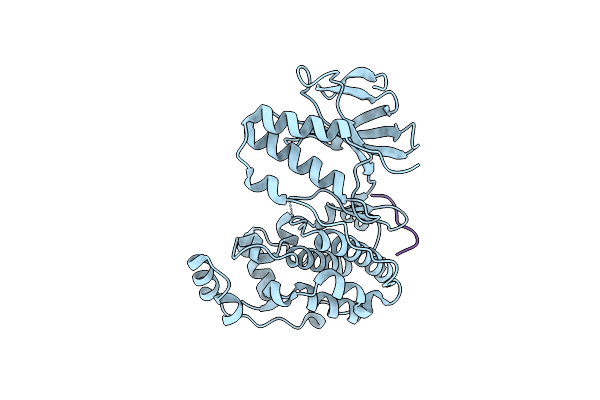 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-19 Classification: TRANSFERASE |
 |
Crystal Structure Of Human Secretory Glutaminyl Cyclase In Complex With The Inhibitor N-(1H-Benzo[D]Imidazol-5-Yl)-1-Phenylmethanesulfonamide (Compound 5)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: ZN, A1D93, GOL, DMS, DMF, PEG |
 |
Crystal Structure Of Human Secretory Glutaminyl Cyclase In Complex With The Inhibitor 3-((2-(1H-Imidazol-5-Yl)Ethyl)Carbamoyl)-4-Amino-1,2,5-Oxadiazole 2-Oxide (Compound 13)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: ZN, A1D94, DMS, GOL, DMF |
 |
Crystal Structure Of Human Golgi Resident Glutaminyl Cyclase In Complex With (Z)-3-((1H-Benzo[D]Imidazol-5-Yl)Methylene)-4-(Piperidin-4-Yloxy)Indolin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D46 |
 |
Crystal Structure Of Human Golgi Resident Glutaminyl Cyclase In Complex With (Z)-3-((1H-Benzo[D]Imidazol-5-Yl)Methylene)-4-((Tetrahydro-2H-Pyran-4-Yl)Oxy)Indolin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.54 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D47 |
 |
Crystal Structure Of Human Secretory Glutaminyl Cyclase In Complex With (S)-1-(1H-Benzo[D]Imidazol-5-Yl)-5-(4-Propoxyphenyl)Imidazolidin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D49 |
 |
Crystal Structure Of Human Secretory Glutaminyl Cyclase In Complex With (Z)-3-((1H-Benzo[D]Imidazol-5-Yl)Methylene)-4-Hydroxyindolin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D48 |
 |
Crystal Structure Of Human Golgi Resident Glutaminyl Cyclase In Complex With (R,Z)-3-((1H-Benzo[D]Imidazol-5-Yl)Methylene)-4-((1-Acetylpyrrolidin-3-Yl)Oxy)Indolin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-06-12 Classification: TRANSFERASE Ligands: ZN, A1D5C |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-05-31 Classification: LIPID TRANSPORT Ligands: PX6, NI |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-04-19 Classification: LIPID TRANSPORT Ligands: D21, NI, CL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-02-08 Classification: TRANSFERASE |
 |
Crystal Structure Of Plasmodium Falciparum Hexose Transporter Pfht1 Bound With Glucose
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-09-09 Classification: TRANSPORT PROTEIN Ligands: BGC, BNG |
 |
Crystal Structure Of Plasmodium Falciparum Hexose Transporter Pfht1 Bound With C3361
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2020-09-09 Classification: TRANSPORT PROTEIN Ligands: F00 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-08-26 Classification: HYDROLASE Ligands: IOD, FLC, PEG |

