Search Count: 30
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: ELECTRON MICROSCOPY Release Date: 2016-12-14 Classification: PROTEIN TRANSPORT |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2016-10-05 Classification: TRANSPORT PROTEIN Ligands: ZN |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2016-10-05 Classification: TRANSPORT PROTEIN Ligands: ZN |
 |
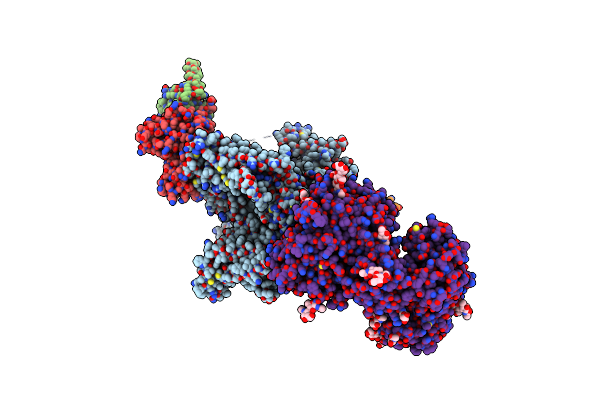
Structure Of The Mammalian Voltage-Gated Calcium Channel Cav1.1 Complex At Near Atomic Resolution
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2016-09-14 Classification: MEMBRANE PROTEIN Ligands: NAG, PC1, CA |
 |
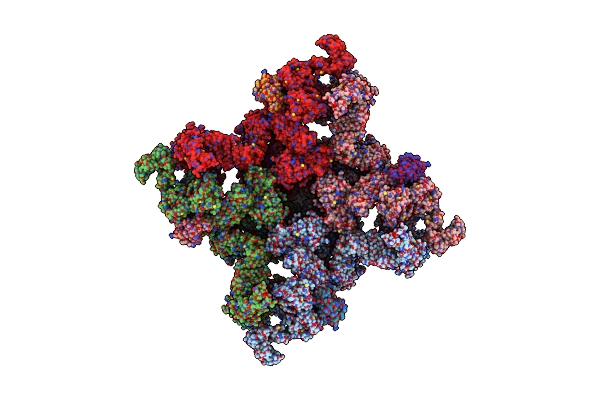
Structure Of The Mammalian Voltage-Gated Calcium Channel Cav1.1 Complex For Classii Map
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2016-09-14 Classification: MEMBRANE PROTEIN Ligands: NAG, CA |
 |
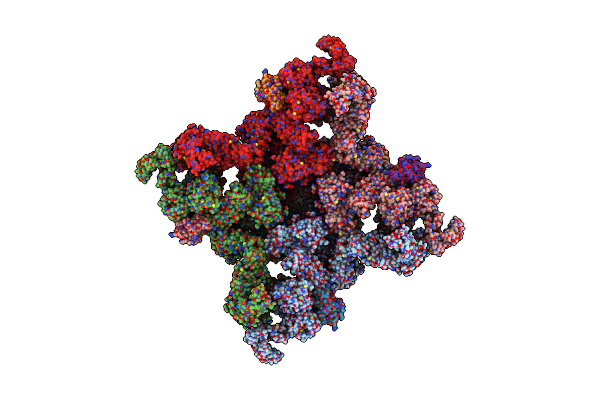
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2016-08-24 Classification: TRANSPORT PROTEIN/ISOMERASE Ligands: ZN |
 |
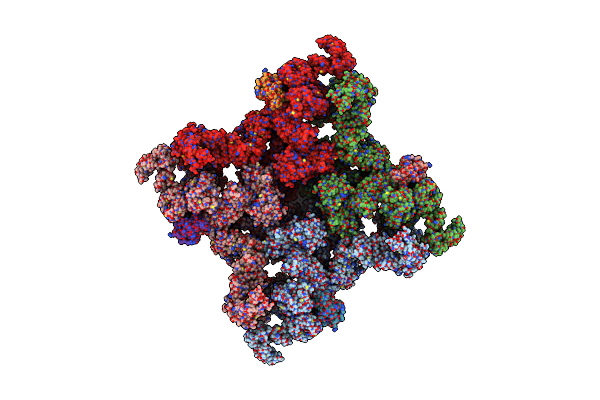
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2016-08-24 Classification: TRANSPORT PROTEIN/ISOMERASE Ligands: ZN |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2016-08-24 Classification: TRANSPORT PROTEIN/ISOMERASE Ligands: ZN |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2016-08-24 Classification: TRANSPORT PROTEIN/ISOMERASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2016-06-15 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2016-06-01 Classification: MEMBRANE PROTEIN Ligands: NAG, CLR |
 |
Organism: Homo sapiens, Hepatitis b virus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-02-10 Classification: APOPTOSIS |
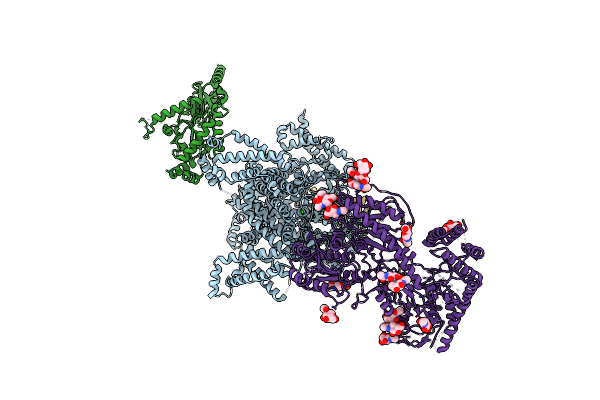 |
Cryo-Em Structure Of The Rabbit Voltage-Gated Calcium Channel Cav1.1 Complex At 4.2 Angstrom
Organism: Rattus norvegicus, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2015-12-30 Classification: MEMBRANE PROTEIN Ligands: CA, NAG, BMA |
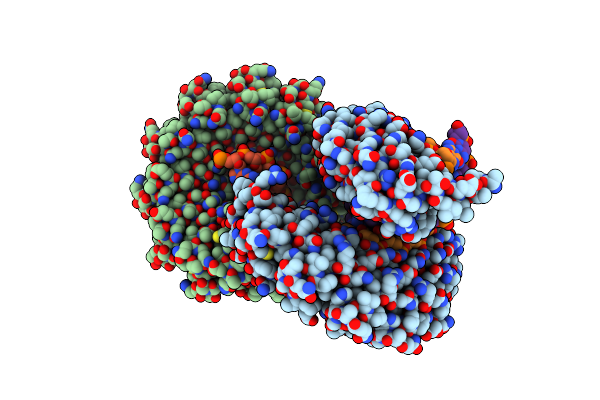 |
Crystal Structure Of The Tal Effector Dhax3 With Ni Rvd At 2.2 Angstrom Resolution
Organism: Xanthomonas campestris pv. armoraciae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-05-28 Classification: DNA binding protein/DNA |
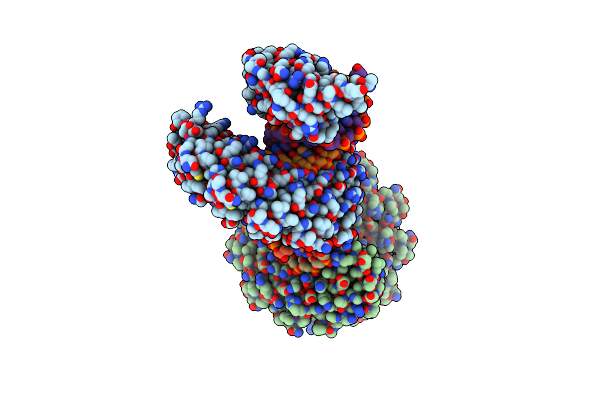 |
Crystal Structure Of The Tal Effector Dhax3 With Ni Rvd At 2.8 Angstrom Resolution
Organism: Xanthomonas campestris pv. armoraciae
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2014-05-28 Classification: DNA binding protein/DNA |
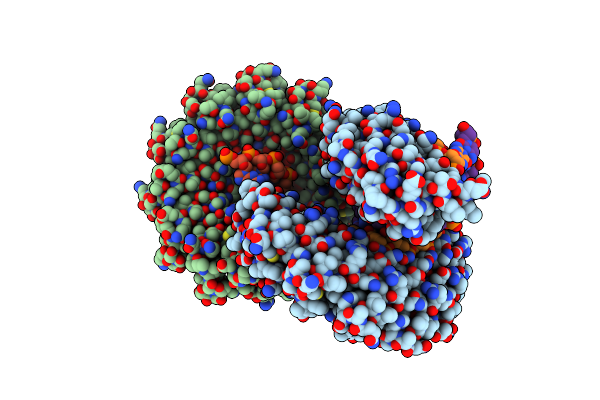 |
Crystal Structure Of Tal Effector Reveals The Recognition Between Asparagine And Adenine
Organism: Xanthomonas campestris pv. armoraciae
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2014-05-28 Classification: DNA binding protein/DNA |
 |
Crystal Structure Of Tal Effector Reveals The Recognition Between Asparagine And Guanine
Organism: Xanthomonas campestris pv. armoraciae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-05-28 Classification: DNA binding protein/DNA |
 |
Crystal Structure Of Tal Effector Reveals The Recognition Between Histidine And Guanine
Organism: Xanthomonas campestris pv. armoraciae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2014-05-28 Classification: DNA binding protein/DNA Ligands: MG |
 |
Organism: Xanthomonas campestris pv. armoraciae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2014-05-28 Classification: DNA binding protein/DNA |
 |
Organism: Xanthomonas campestris pv. armoraciae
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-05-28 Classification: DNA binding protein/DNA Ligands: MG |

