Search Count: 1,472
 |
Organism: Homo sapiens
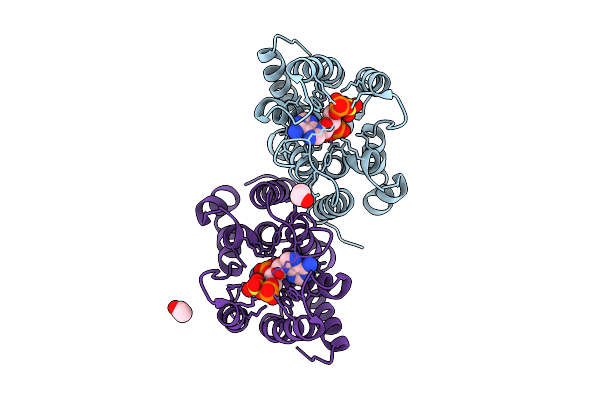
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: ZN, PPS, EDO, ACY |
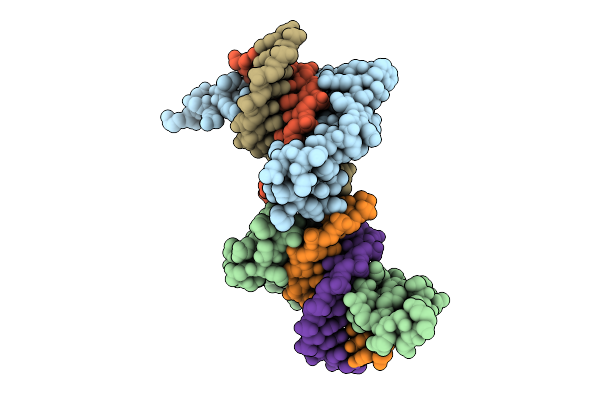 |
Crystal Structure Of The First Three Zinc Finger Domains Of Zbtb20 In Complex With Dna
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
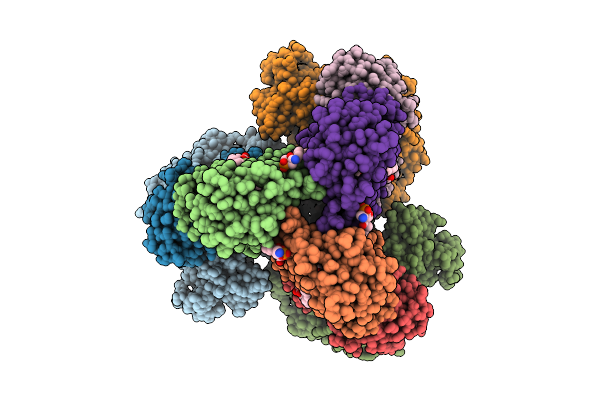 |
Cryo-Em Structure Of The Apo-Form Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
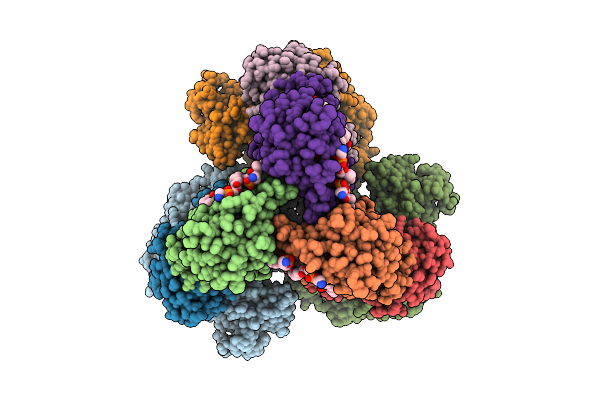 |
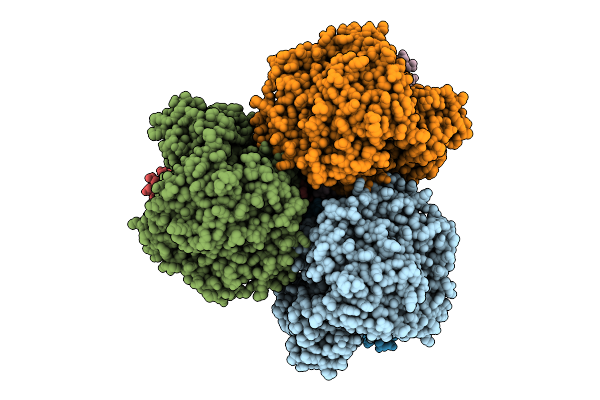
Cryo-Em Structure Of The Lipid-Bound Succiante Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
 |
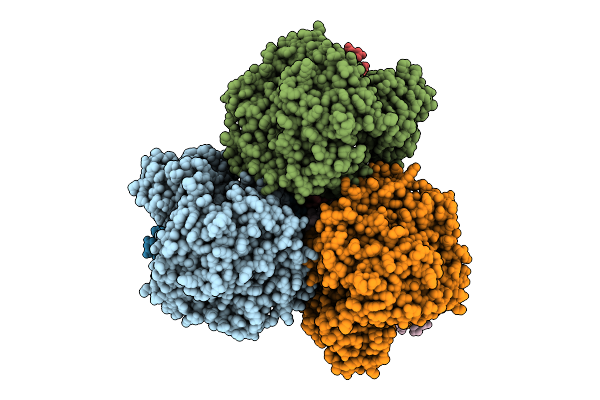
Cryo-Em Structure Of The Mk7-Bound Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SF4, FES, F3S, HEM, LMT, MQ7 |
 |
Cryo-Em Structure Of The Mk4-Bound Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SF4, FES, F3S, HEM, LMT, 1L3 |
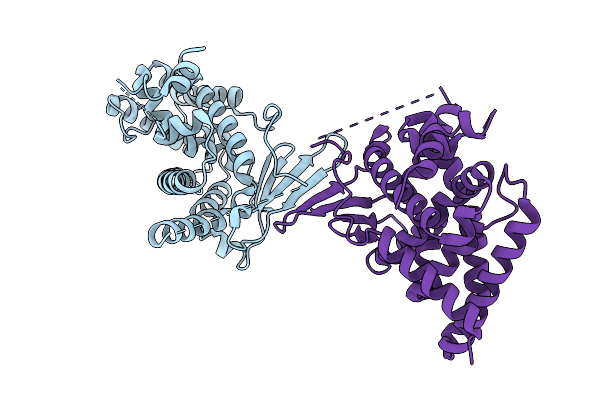 |
Crystal Structure Of The L411A Mutant Of Pregnane X Receptor Ligand Binding Domain (Apo Form)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSCRIPTION |
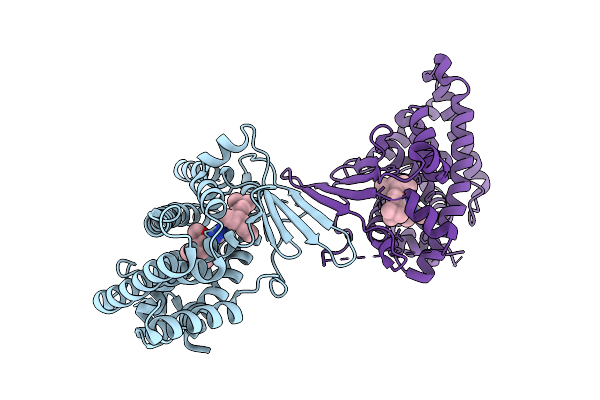 |
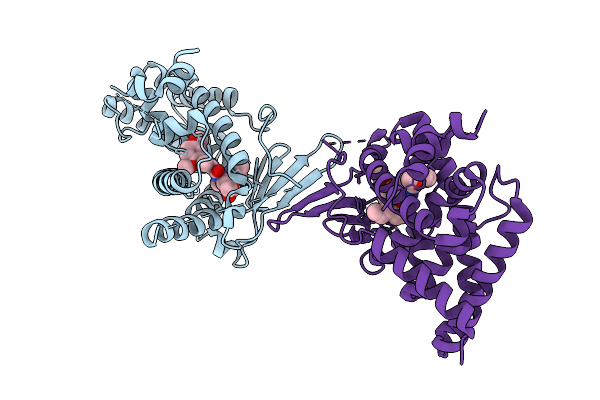
Crystal Structure Of The L411A Mutant Of Pregnane X Receptor Ligand Binding Domain In Complex With Sjpyt-328
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: WU2 |
 |
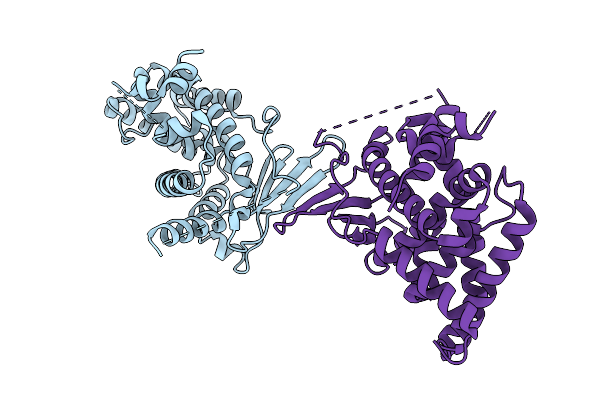
Crystal Structure Of The L411A Mutant Of Pregnane X Receptor Ligand Binding Domain In Complex With Sjpyt-331
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: WU6 |
 |
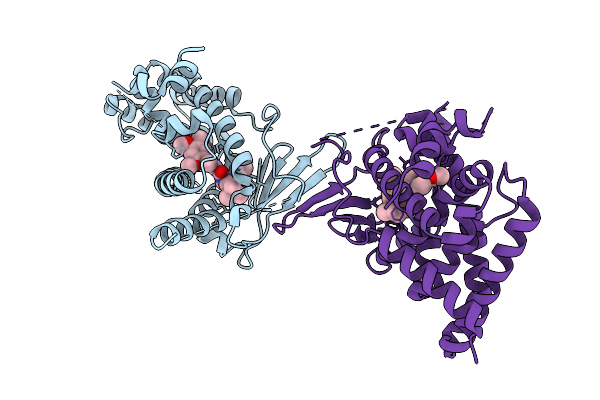
Crystal Structure Of The L411W Mutant Of Pregnane X Receptor Ligand Binding Domain (Apo Form)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSCRIPTION |
 |
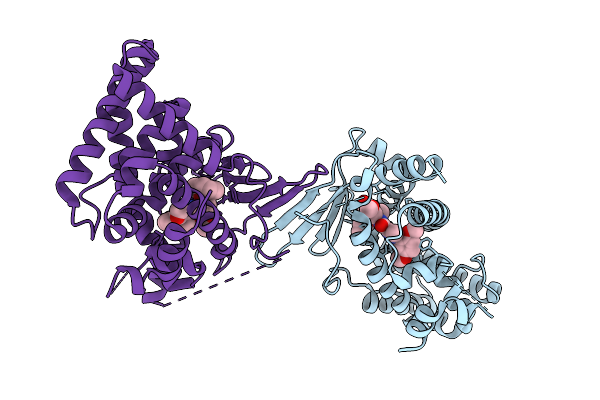
Crystal Structure Of The L411W Mutant Of Pregnane X Receptor Ligand Binding Domain In Complex With Sjpyt-328
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: WU2 |
 |
Crystal Structure Of The L411W Mutant Of Pregnane X Receptor Ligand Binding Domain In Complex With Sjpyt-331
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: WU6 |
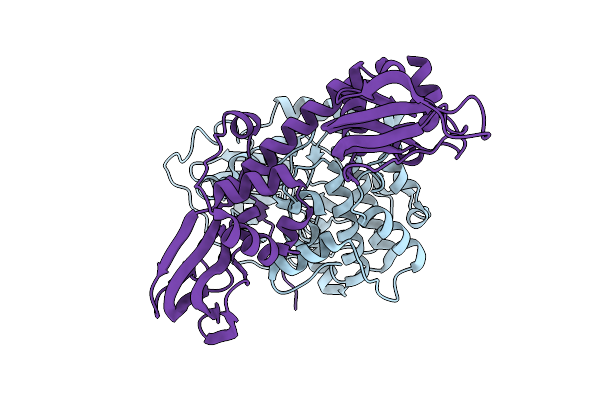 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: OT5 |
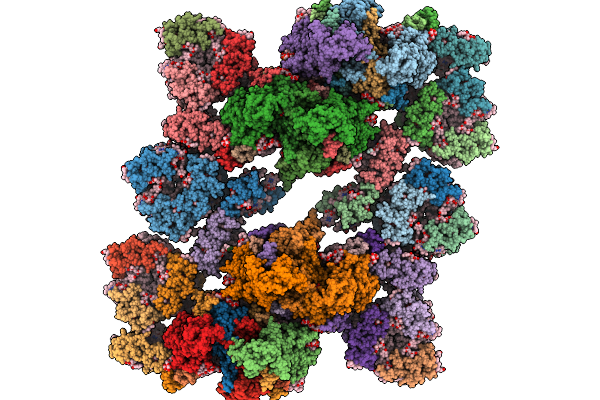 |
Coordinates Of Cryo-Em Structure Of The Arabidopsis Thaliana C4S4M4-Type Psii Supercomplex
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: PHOTOSYNTHESIS Ligands: CHL, CLA, LUT, NEX, LHG, XAT, BCR, LMG, PL9, SQD, HEM, DGD, BCT, OEX, FE2, PHO |
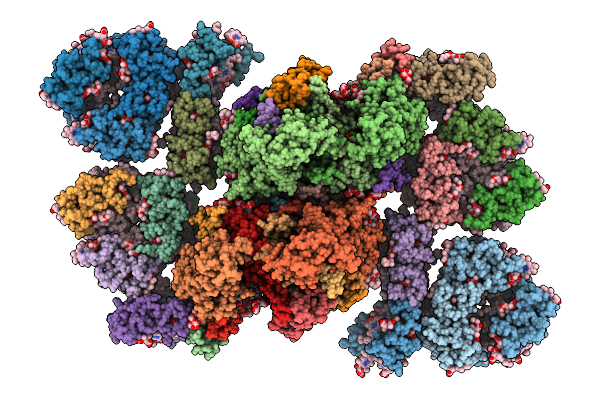 |
Coordinates Of Cryo-Em Structure Of The Arabidopsis Thaliana C2S2M2-Type Psii Supercomplex
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: PHOTOSYNTHESIS Ligands: CHL, CLA, LUT, NEX, XAT, LHG, BCR, OEX, FE2, PHO, LMG, SQD, DGD, BCT, PL9, HEM |
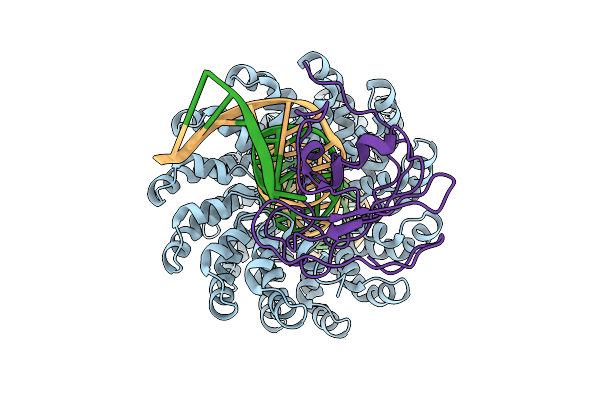 |
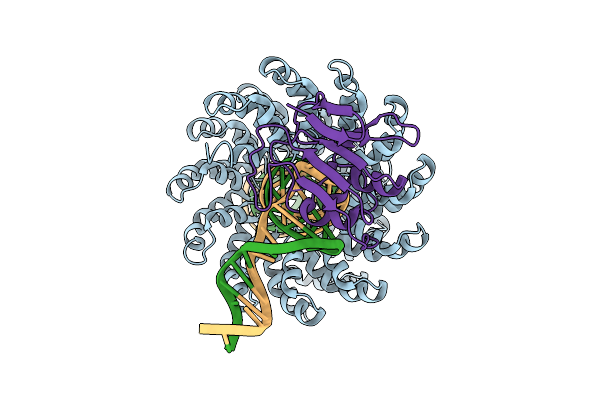
Organism: Xanthomonas, Burkholderia cenocepacia h111, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
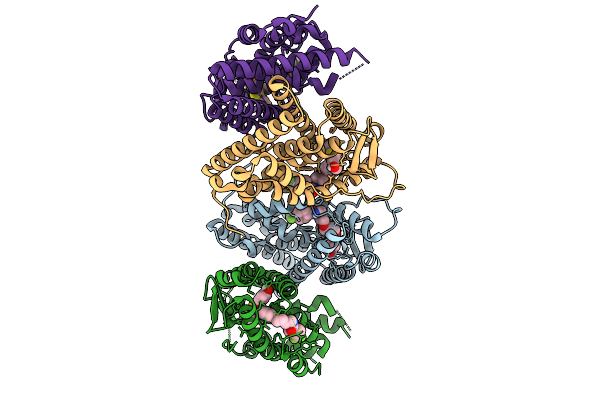
Organism: Xanthomonas, Burkholderia cenocepacia h111, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
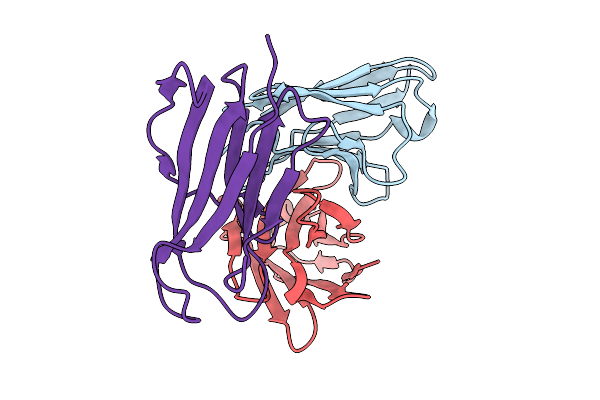 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Na98
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: NUCLEAR PROTEIN Ligands: A1AIZ |

