Search Count: 268
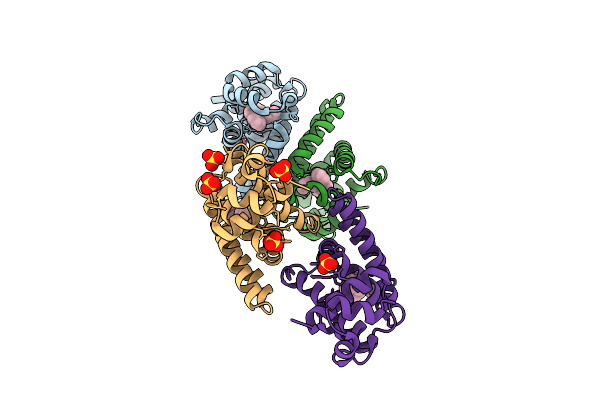 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. 14028s
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: PHOTOSYNTHESIS Ligands: A1EKZ, SO4 |
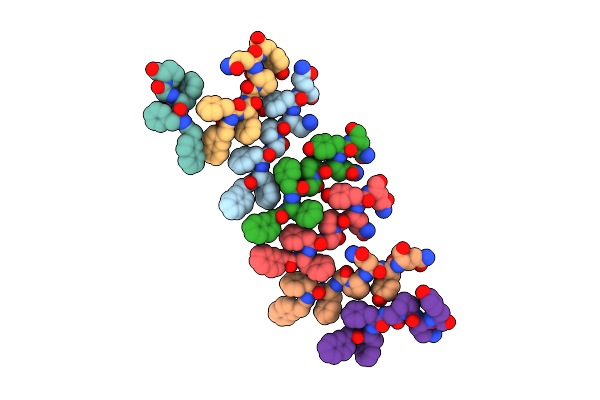 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: PROTEIN FIBRIL |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: PROTEIN FIBRIL |
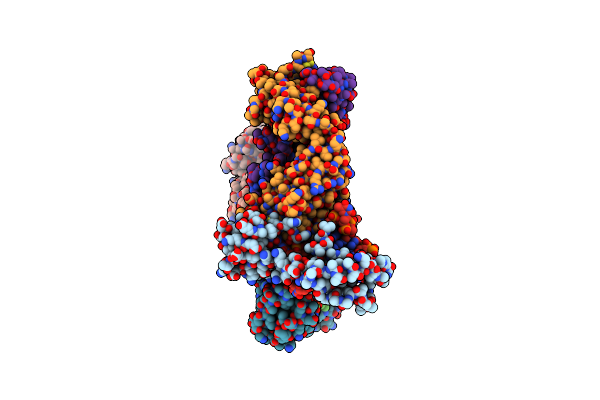 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-04-30 Classification: TRANSCRIPTION/DNA Ligands: K |
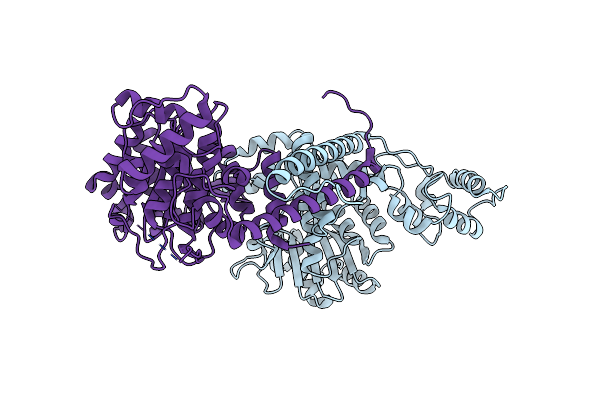 |
Organism: Monkeypox virus zaire-96-i-16
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: VIRAL PROTEIN |
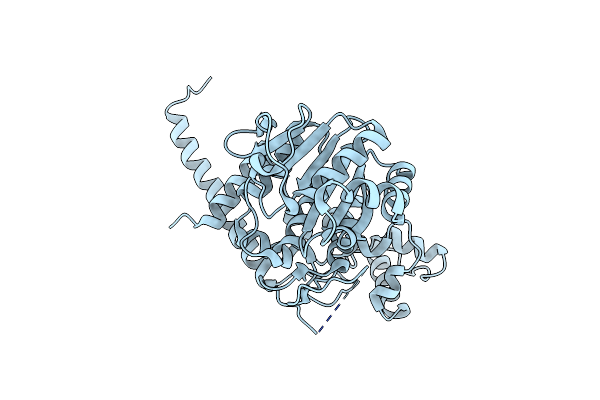 |
Crystal Structure Of The Monkeypox Virus N-Tagged I7L Protease In The I4 Space Group
Organism: Monkeypox virus (strain zaire-96-i-16)
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: VIRAL PROTEIN |
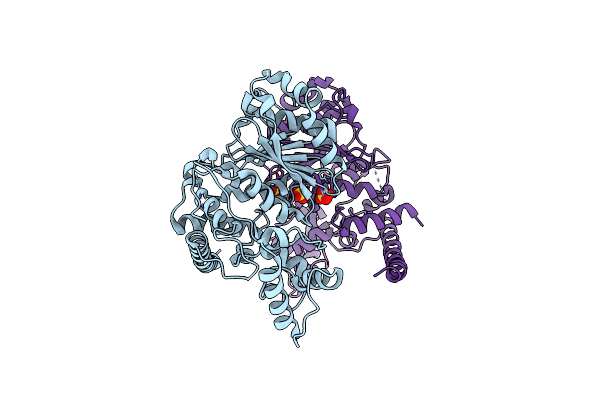 |
Crystal Structure Of The Monkeypox Virus N-Tagged I7L Protease In The P212121 Space Group
Organism: Monkeypox virus (strain zaire-96-i-16)
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: VIRAL PROTEIN Ligands: SO4 |
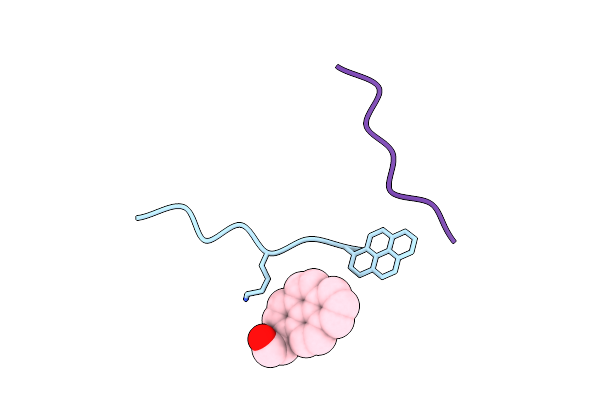 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN FIBRIL Ligands: OG9 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: TRANSFERASE Ligands: SAH, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-15 Classification: TRANSFERASE Ligands: GOL, SAH, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-01-15 Classification: TRANSFERASE Ligands: SAH, ZN, PEG, PG4, EOH, GOL |
 |
Molecular Basis Of The Phosphorothioation-Sensing Antiphage Defence System Dndbcde-Dndi
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN Ligands: PGE, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
Cryo-Em Structure Of Fpgalactosaminidase From Flavonifractor Plautii In Apo State
Organism: Flavonifractor plautii
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: HYDROLASE |
 |
Macrocyclic Inhibitors Targeting The Prime Site Of The Fibrinolytic Serine Protease Plasmin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-30 Classification: HYDROLASE/INHIBITOR Ligands: A1AHR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/HYDROLASE/IMMUNE SYSTEM Ligands: NAG, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/HYDROLASE/IMMUNE SYSTEM Ligands: NAG, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |

