Search Count: 154
 |
Crystal Structure Of Crimean-Congo Hemorrhagic Fever Virus Cap-Snatching Endonuclease
Organism: Orthonairovirus haemorrhagiae, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Manganese Ions
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E668A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E682A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
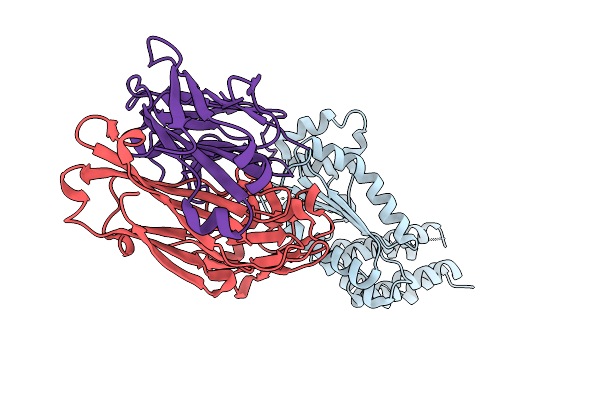 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (D719A Mutant)
Organism: Mus musculus, Kasokero virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With 2,4-Dioxo-4-Phenylbutanoic Acid (Dpba)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, XI7 |
 |
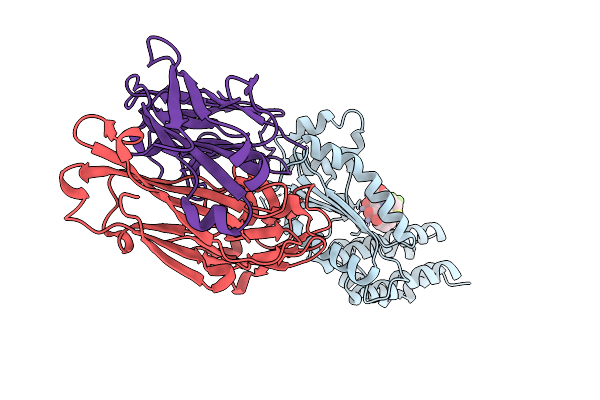
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With L-742,001
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, 0N8 |
 |
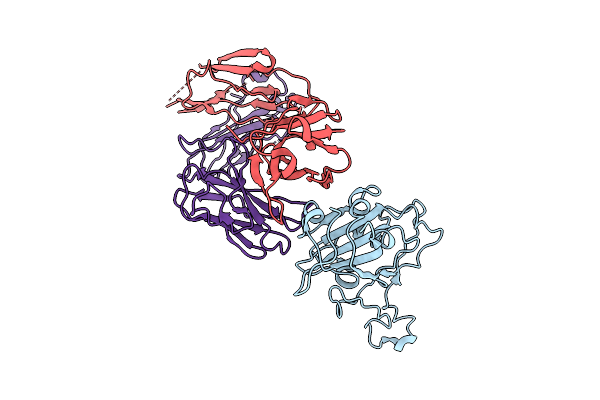
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Baloxavir Acid (Bxa)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, E4Z |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
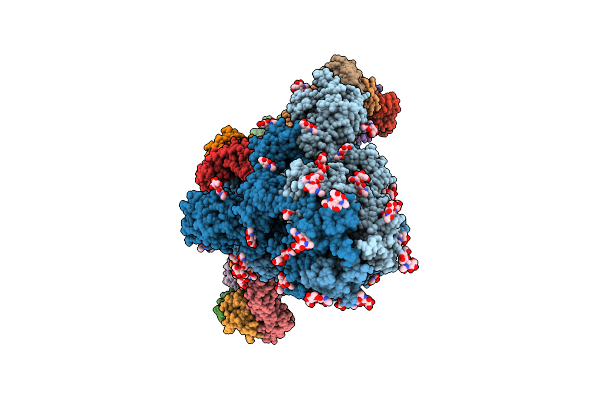
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
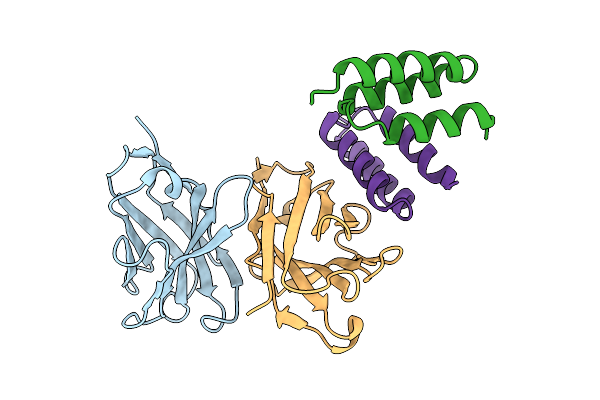
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
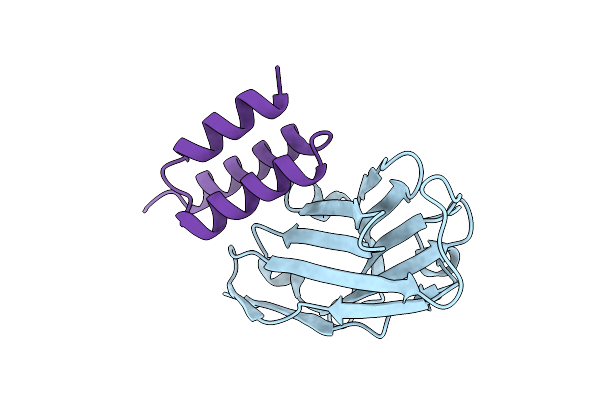
Crystal Structure Of S. Aureus Protein A Bound To A Human Single-Domain Antibody
Organism: Homo sapiens, Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
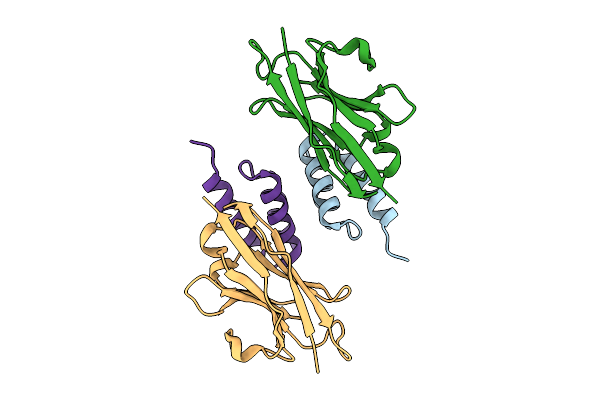
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Camelidae, Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Camelus dromedarius
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
Organism: Streptomyces coelicolor a3(2)
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: OXIDOREDUCTASE Ligands: CU, RU |

