Search Count: 189
 |
Open State Without Nuqm And Without Flavoprotein (Classification State 4) Of Pichia Pastoris Mitochondrial Complex I In Cmsp26 Nanodiscs
Organism: Komagataella pastoris
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: SF4, PLC, K, 3PE, CDL, NDP, ZN, EHZ |
 |
Open State Without Nuqm And With Flavoprotein (Classification State 3) Of Pichia Pastoris Mitochondrial Complex I In Cmsp26 Nanodiscs
Organism: Komagataella pastoris
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: SF4, PLC, FES, FMN, K, 3PE, CDL, NDP, ZN, EHZ |
 |
Closed State With Nuqm And Without Flavoprotein (Classification State 2) Of Pichia Pastoris Mitochondrial Complex I In Cmsp26 Nanodiscs
Organism: Komagataella pastoris
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: PLC, SF4, FES, K, 3PE, CDL, NDP, ZN, EHZ |
 |
Closed State With Nuqm And With Flavoprotein (Classification State 1) Of Pichia Pastoris Mitochondrial Complex I In Cmsp26 Nanodiscs
Organism: Komagataella pastoris
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: PLC, SF4, FES, FMN, K, 3PE, CDL, NDP, ZN, EHZ |
 |
Cryo-Em Structure Of Halothiobacillus Neapolitanus Alpha-Carboxysome T=4 Mini-Shell Containing Ctd Truncated Mutant Of Csosca
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Alpha-Carboxysome T=4 Mini-Shell Containing Ctd Only Mutant Of Csosca
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: STRUCTURAL PROTEIN |
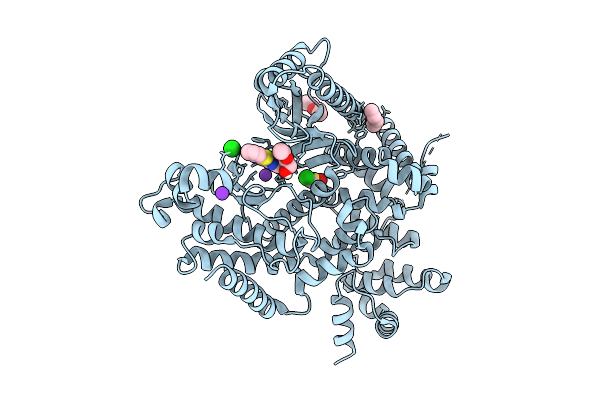 |
The Structure Of Human Vacuolar Protein Sorting 34 Catalytic Domain Bound To Rd-I-53
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: TRANSFERASE Ligands: A1A6F, ETX, ETZ, DMS, K, CL |
 |
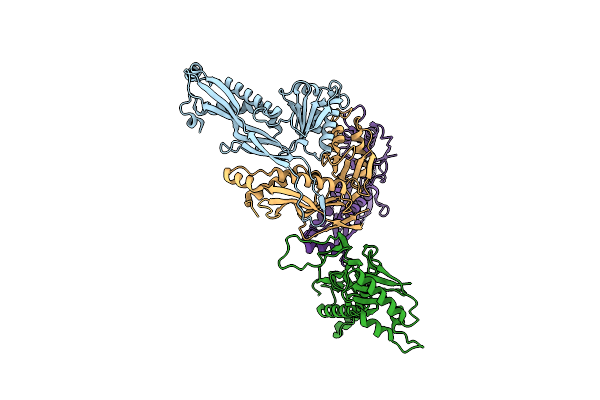
Beta Carbonic Anhydrase Csosca From The Halothiobacillus Neapolitanus Alpha-Carboxysome
Organism: Streptococcus sp. 'group g', Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LYASE Ligands: ZN |
 |
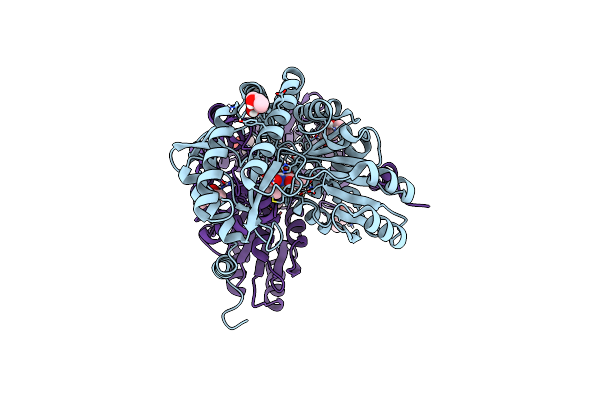
Single Particle Cryo-Em Reconstruction Of Bacillus Methanolicus Encapsulin Nano Compartment
Organism: Bacillus methanolicus mga3
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: STRUCTURAL PROTEIN |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: LIGASE Ligands: MPD, A1H4V |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell:Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 19)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 16)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 9)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T=9 Q=12)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Midi-Shell: Icosahedral Assembly From Csos4A/4B/1A/1B/1C/1D And Csos2 C-Terminal Co-Expression (T = 13)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Organism: Halothiobacillus neapolitanus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN |
 |
Cryo-Em Structure Of Carboxysomal Mini-Shell Icosahedral Assembly From Co-Expression Of Csos1C, Csos4A, And Csos2-C (T = 9)
Organism: Halothiobacillus neapolitanus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: STRUCTURAL PROTEIN Ligands: MG |
 |
The Structure Of Human Vacuolar Protein Sorting 34 Catalytic Domain Bound To Mes
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-11-06 Classification: TRANSFERASE Ligands: MES, PG4, DMS, PEG, GOL, CL, K |
 |
Crystal Structure Of A Mes Bound Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-08-28 Classification: TRANSPORT PROTEIN Ligands: MES |
 |
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-08-28 Classification: TRANSPORT PROTEIN Ligands: EPE, EDO |

