Search Count: 30
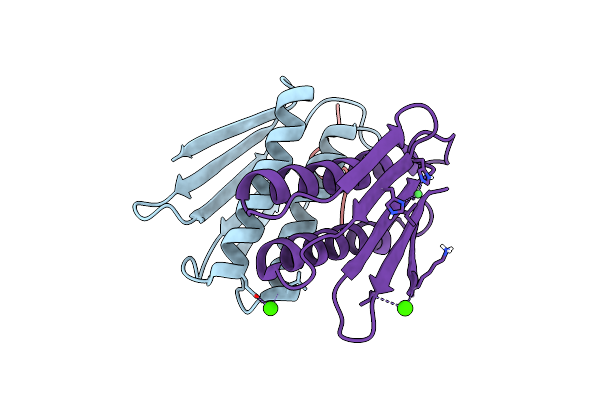 |
Crystal Structure Of An Essential Ribosomal Processing Protease Prp From S. Aureus In Complex With A Substrate Peptide
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: CA, NI |
 |
Crystal Structure Of An Essential Ribosomal Processing Protease Prp From S. Aureus In Complex With A Covalently Linked Product Peptide
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: CA |
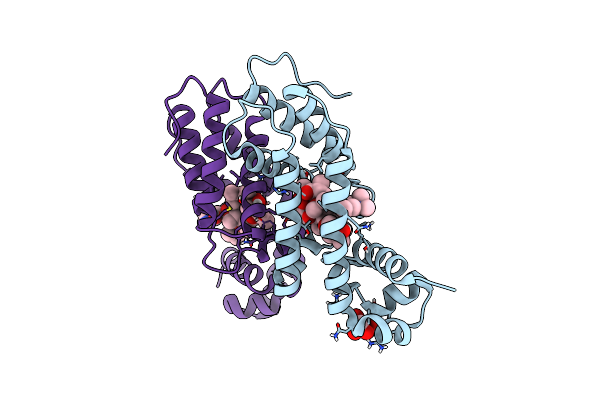 |
Directed Evolution Of A Biosensor Selective For The Macrolide Antibiotic Clarithromycin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-19 Classification: DNA BINDING PROTEIN/ANTIBIOTIC Ligands: CTY, FLC |
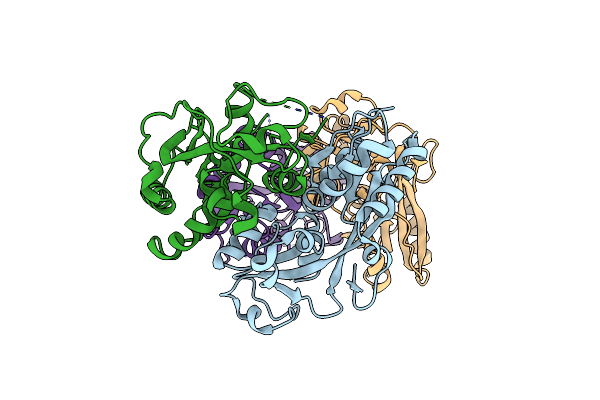 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:4.15 Å Release Date: 2013-07-31 Classification: TRANSCRIPTION REGULATOR |
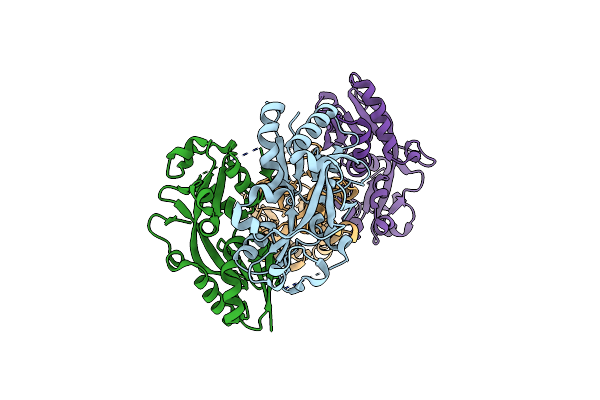 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-04-17 Classification: TRANSCRIPTION REGULATOR |
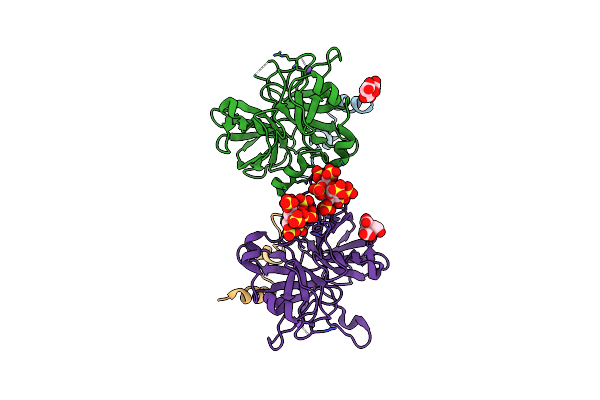 |
2.2 Angstrom Crystal Structure Of The Complex Between Bovine Thrombin And Sucrose Octasulfate
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-07-20 Classification: Hydrolase/Hydrolase Inhibitor |
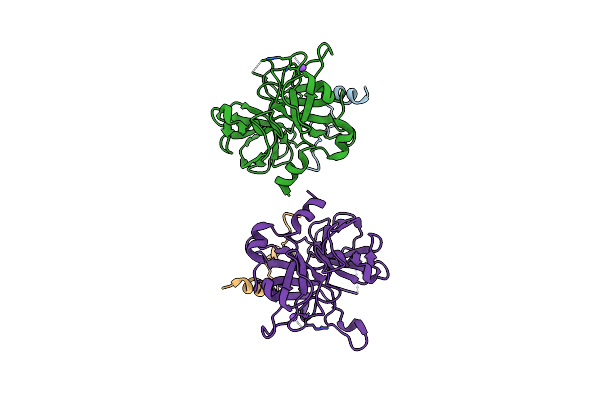 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2011-07-20 Classification: HYDROLASE |
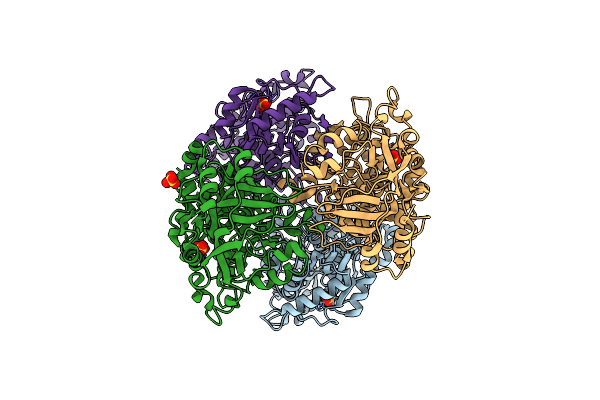 |
2.4 Angstrom Crystal Structure Of Streptomyces Collinus Crotonyl Coa Carboxylase/Reductase
Organism: Streptomyces collinus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-07-21 Classification: OXIDOREDUCTASE Ligands: SO4 |
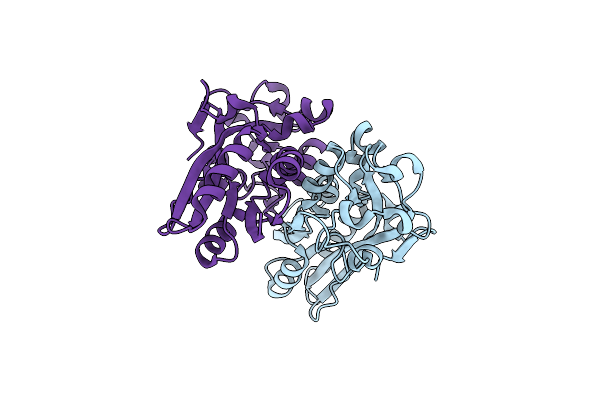 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2010-06-09 Classification: TRANSCRIPTION |
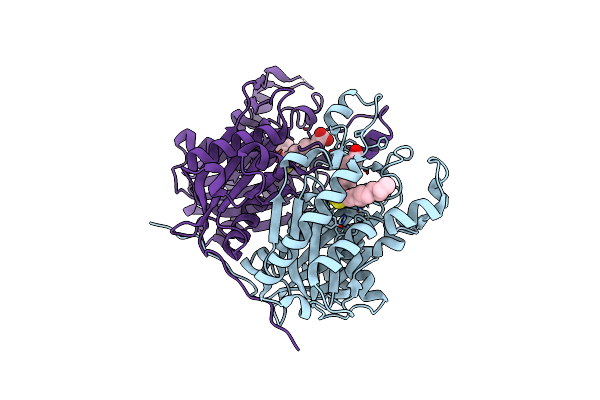 |
Crystal Structure Of The Complex Between The Mycobacterium Beta-Ketoacyl-Acyl Carrier Protein Synthase Iii (Fabh) And 11-[(Decyloxycarbonyl)Dithio]-Undecanoic Acid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-05-06 Classification: TRANSFERASE Ligands: UDT, MDX |
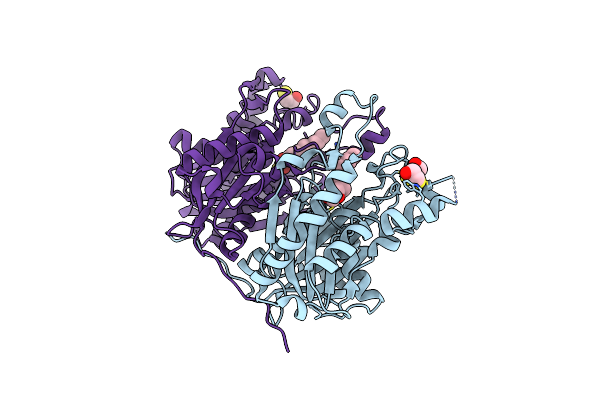 |
Crystal Structure Of The Complex Between The A246F Mutant Of Mycobacterium Beta-Ketoacyl-Acyl Carrier Protein Synthase Iii (Fabh) And Ss-(2-Hydroxyethyl) O-Decyl Ester Carbono(Dithioperoxoic) Acid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2008-05-06 Classification: TRANSFERASE Ligands: DFD, BME |
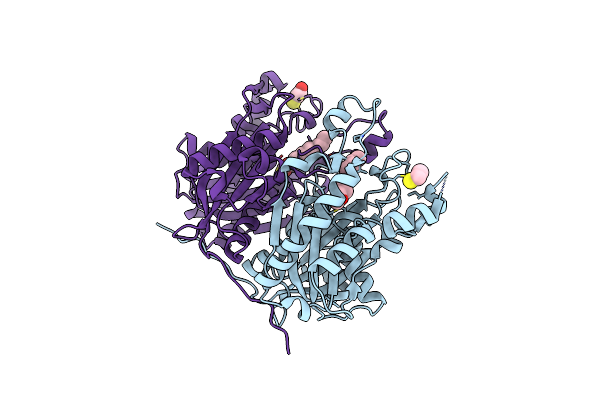 |
Crystal Structure Of The Complex Between The Mycobacterium Beta-Ketoacyl-Acyl Carrier Protein Synthase Iii (Fabh) And Ss-(2-Hydroxyethyl)-O-Decyl Ester Carbono(Dithioperoxoic) Acid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-05-06 Classification: TRANSFERASE Ligands: DFD, BME |
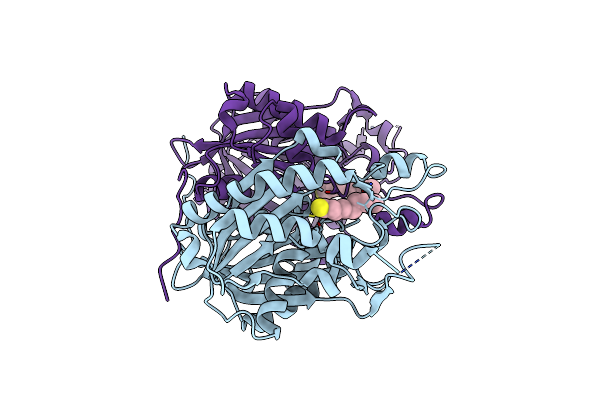 |
Crystal Structure Of The Complex Between The A246F Mutant Of Mycobacterium Beta-Ketoacyl-Acyl Carrier Protein Synthase Iii (Fabh) And 11-(Decyldithiocarbonyloxy)-Undecanoic Acid
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-05-06 Classification: TRANSFERASE Ligands: D1T |
 |
2.6 Angstrom Crystal Structure Of The Complex Between 11-(Decyldithiocarbonyloxy)-Undecanoic Acid And Mycobacterium Tuberculosis Fabh.
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-05-06 Classification: TRANSFERASE Ligands: VZZ, D1T |
 |
Methanethiol-Cys 112 Inhibition Complex Of E. Coli Ketoacyl Synthase Iii (Fabh) And Coenzyme A (High Concentration (1.7Mm) Soak)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-06-12 Classification: TRANSFERASE Ligands: SO4, COA, MEE |
 |
Methanethiol-Cys 112 Inhibition Complex Of E. Coli Ketoacyl Synthase Iii (Fabh) And Coenzyme A
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-06-05 Classification: TRANSFERASE Ligands: COA, MEE, SO4 |
 |
2.7 Angstrom Crystal Structure Of The Complex Between The Nuclear Snorna Decapping Nudix Hydrolase X29 And Manganese
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2006-03-28 Classification: HYDROLASE Ligands: MN |
 |
2.6 Angstrom Crystal Structure Of The Complex Between The Nuclear Snorna Decapping Nudix Hydrolase X29 And Manganese In The Presence Of 7-Methyl-Gdp
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-03-28 Classification: TRANSLATION,HYDROLASE Ligands: MN, POP |
 |
2.45 Angstrom Crystal Structure Of The Complex Between The Nuclear Snorna Decapping Nudix Hydrolase X29 And Manganese In The Presence Of 7-Methyl-Gtp
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2006-03-28 Classification: TRANSLATION,HYDROLASE Ligands: MN, POP |
 |
2.45 Angstrom Crystal Structure Of The Complex Between The Nuclear Snorna Decapping Nudix Hydrolase X29, Manganese And Gtp
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2006-03-28 Classification: TRANSLATION,HYDROLASE Ligands: MN, GTP |

