Search Count: 102
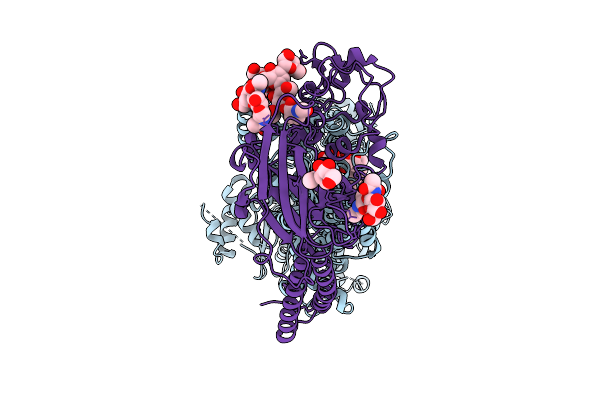 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
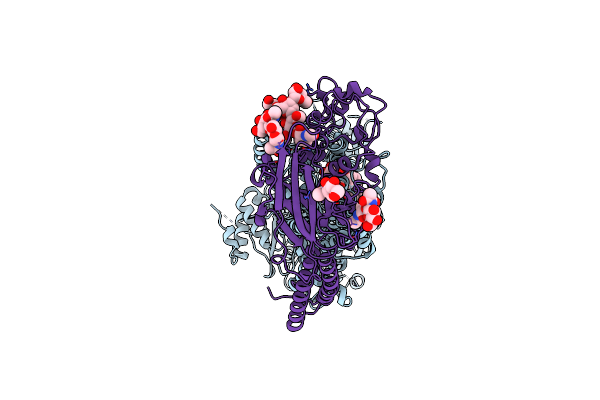 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
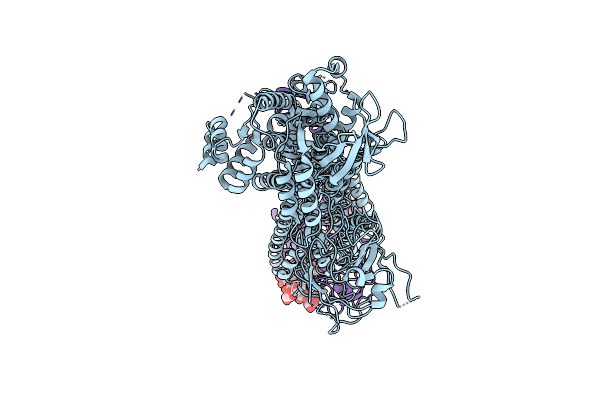 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 In The E1 State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: LIPID TRANSPORT Ligands: NAG, MG |
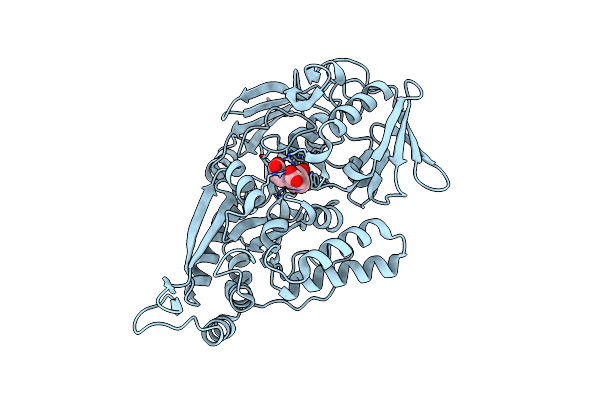 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-04-10 Classification: TRANSPORT PROTEIN Ligands: II1, NI |
 |
Candida Albicans Glutaminyl Trna Synthetase (Gln4) In Complex With N-Pyrimidinyl-Beta-Thiophenylacrylamide
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2024-03-06 Classification: LIGASE/LIGASE INHIBITOR Ligands: ZN, WVK |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2023-04-26 Classification: MEMBRANE PROTEIN Ligands: SO4, BOG, FE, OX8 |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Gentamicin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: LLL, EDO, MG, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Tobramycin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: TOY, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Kanamycin B And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: 9CS, COA, EDO, SO4, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Paromomycin And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: COA, PAR, GOL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Neomycin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: NMY, EDO |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma H135A Mutant, Complex With Tobramycin And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: TOY, EDO, COA |
 |
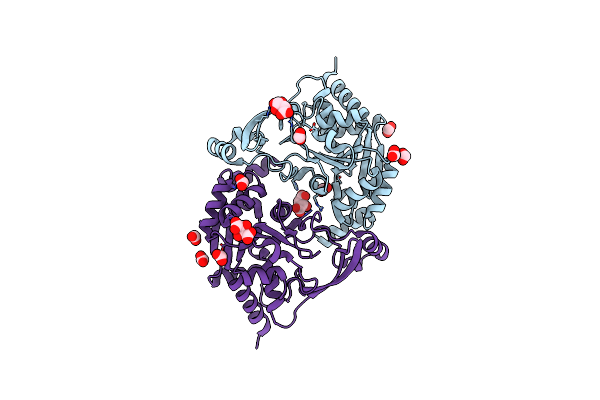
Crystal Structure Of Escherichia Coli Clpp Covalently Inhibited By Clipibicyclene
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-02-02 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: MPD, ACT, ZGV |
 |
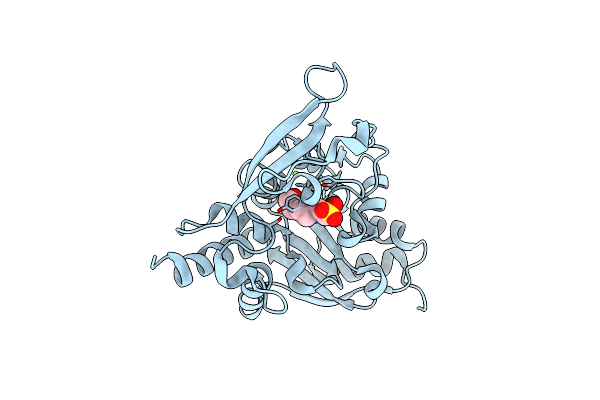
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-02-03 Classification: TRANSFERASE Ligands: TAR, FMT, CL |
 |
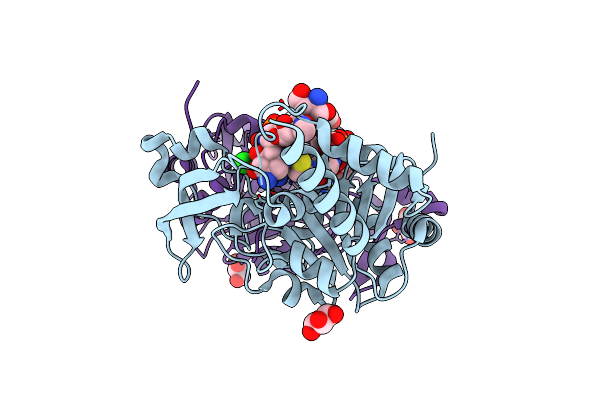
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-01-20 Classification: TRANSFERASE Ligands: EPE, MG |
 |
Crystal Structure Of Meta-Aac0038, An Environmental Aminoglycoside Resistance Enzyme, Mutant H168A In Complex With Apramycin And Coa
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2020-10-21 Classification: TRANSFERASE/ANTIBIOTIC Ligands: GOL, COA, AM2, CL |
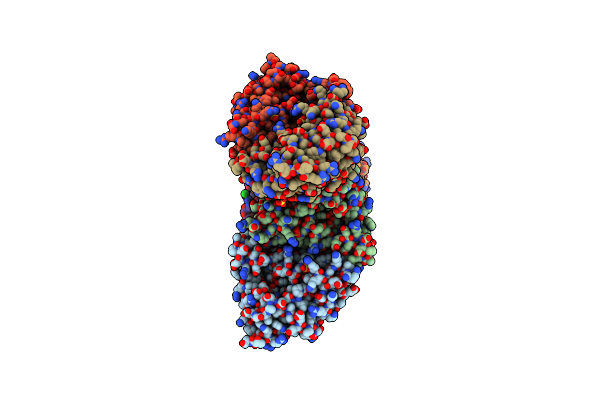 |
Crystal Structure Of Meta-Aac0038, An Environmental Aminoglycoside Resistance Enzyme, H29A Mutant Apoenzyme
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2018-10-24 Classification: TRANSFERASE Ligands: SO4, CL |
 |
Crystal Structure Of Meta-Aac0038, An Environmental Aminoglycoside Resistance Enzyme, H168A Mutant In Complex With Acetyl-Coa
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-10-24 Classification: TRANSFERASE Ligands: ACO, CL, GOL, SO4, PE3 |
 |
Crystal Structure Of Aminoglycoside Acetyltransferase Aac(3)-Iva, Apoenzyme
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-10-24 Classification: TRANSFERASE Ligands: CL, ZN |
 |
Crystal Structure Of Aminoglycoside Acetyltransferase Aac(3)-Iva, H154A Mutant, In Complex With Apramycin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-10-24 Classification: TRANSFERASE/ANTIBIOTIC Ligands: AM2, ZN, EDO, PO4, EPE, GOL |

