Search Count: 188
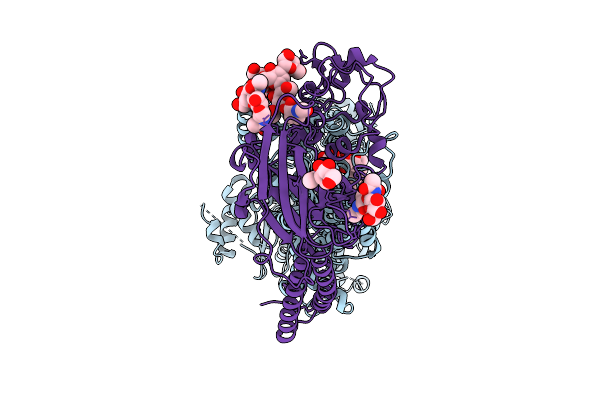 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
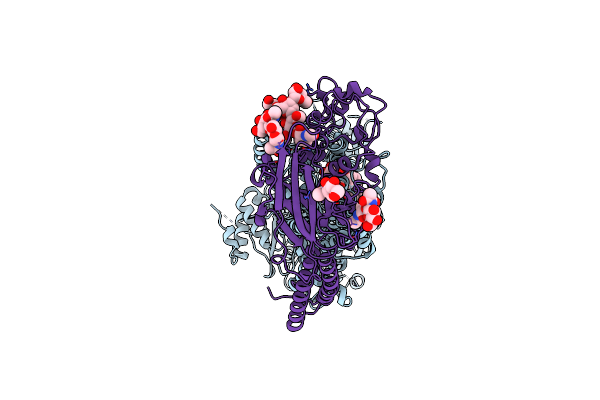 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 Bound With Butyrolactol A In The E2P State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: TRANSLOCASE/INHIBITOR Ligands: NAG, BEF, MG, A1BD6 |
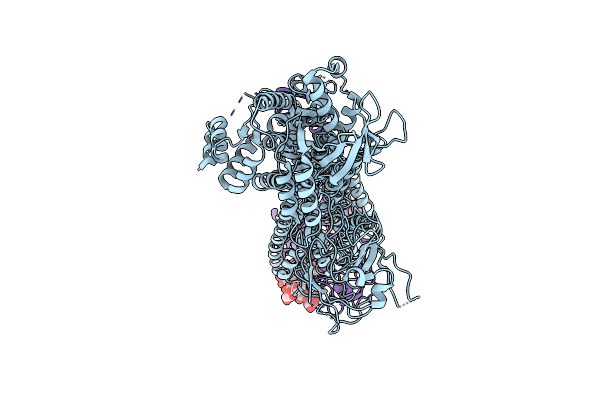 |
Cryo-Em Structure Of The C. Neoformans Lipid Flippase Apt1-Cdc50 In The E1 State
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: LIPID TRANSPORT Ligands: NAG, MG |
 |
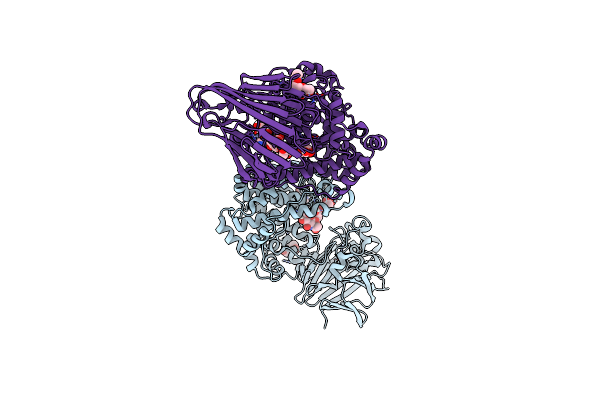
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-12-04 Classification: LYASE Ligands: ZN |
 |
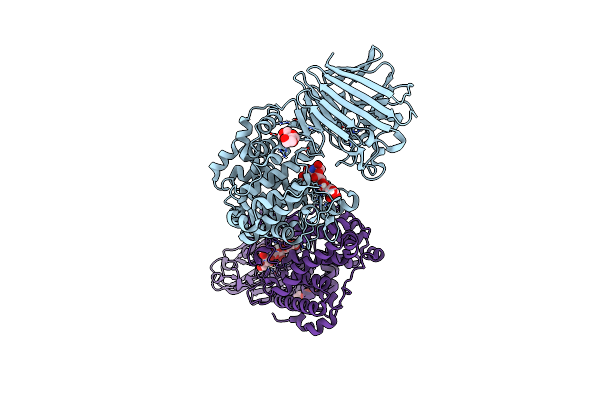
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-04 Classification: LYASE Ligands: P4K, MES, ZN |
 |
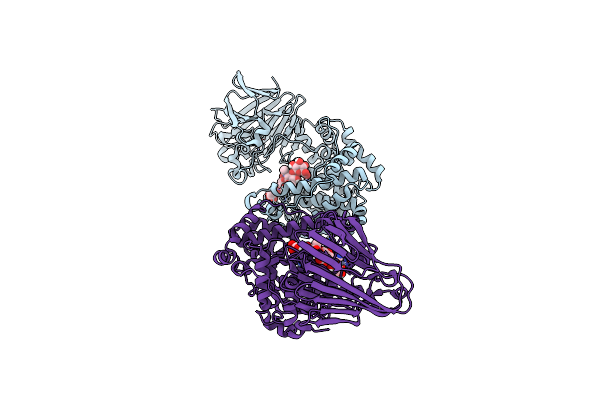
Polysaccharide Lyase Btpl33Ha (Bt4410) Y291A With Hadp4 Collected At 1.22 A
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-12-04 Classification: LYASE Ligands: P4K, ZN |
 |
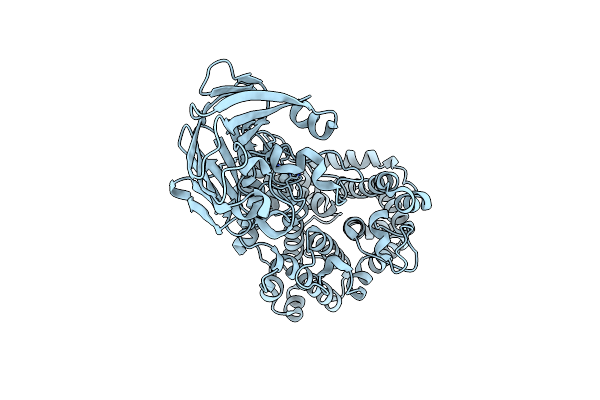
Polysaccharide Lyase Btpl33Ha (Bt4410) Y291A With Ha Dp4 Collected At 1.33 A
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-12-04 Classification: LYASE Ligands: ZN |
 |
Organism: Bacteroides cellulosilyticus wh2
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-12-04 Classification: LYASE Ligands: ZN |
 |
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-12-04 Classification: LYASE Ligands: ZN |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin B, Mrna, Aminoacylated A-Site Phe-Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.60A Resolution
Organism: Escherichia coli, Paenibacillus, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, K, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Lasso Peptide Lariocidin And Protein Y At 2.60A Resolution
Organism: Escherichia coli k-12, Paenibacillus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: RIBOSOME Ligands: MG, ARG, MPD, ZN, SF4 |
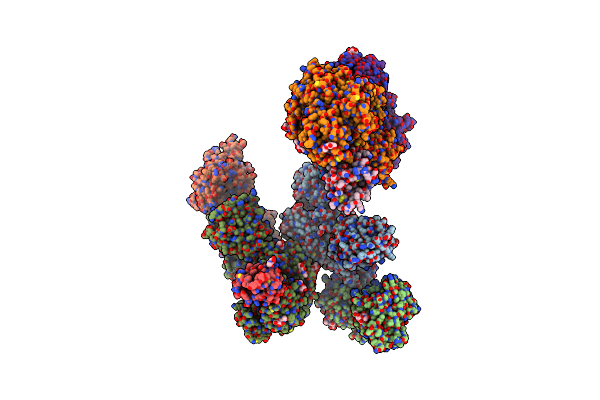 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.70 Å Release Date: 2024-04-24 Classification: SIGNALING PROTEIN Ligands: NAG |
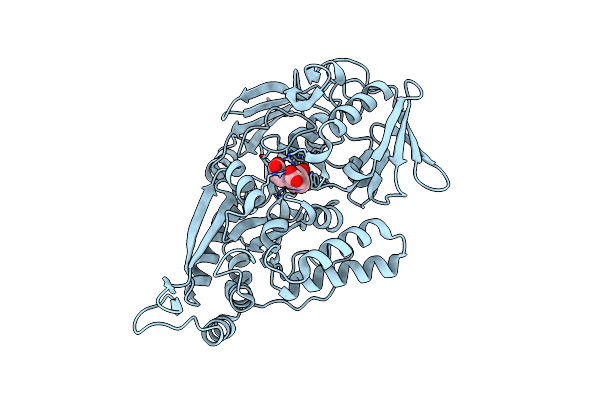 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-04-10 Classification: TRANSPORT PROTEIN Ligands: II1, NI |
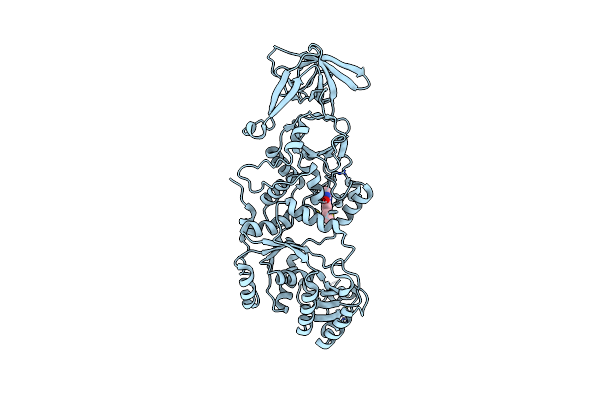 |
Candida Albicans Glutaminyl Trna Synthetase (Gln4) In Complex With N-Pyrimidinyl-Beta-Thiophenylacrylamide
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2024-03-06 Classification: LIGASE/LIGASE INHIBITOR Ligands: ZN, WVK |
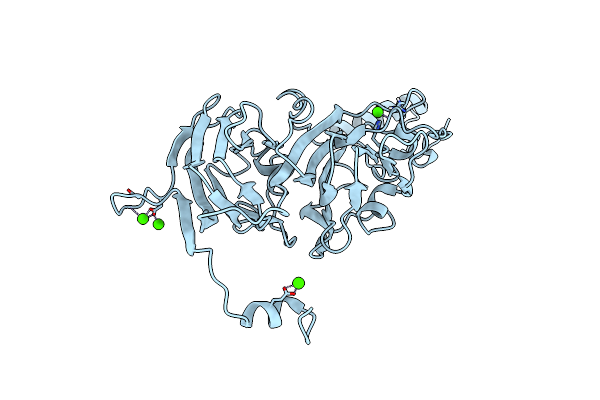 |
Organism: Dysgonomonas gadei
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: CA |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2023-04-26 Classification: MEMBRANE PROTEIN Ligands: SO4, BOG, FE, OX8 |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Gentamicin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: LLL, EDO, MG, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Tobramycin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: TOY, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Kanamycin B And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: 9CS, COA, EDO, SO4, CL |

