Search Count: 29
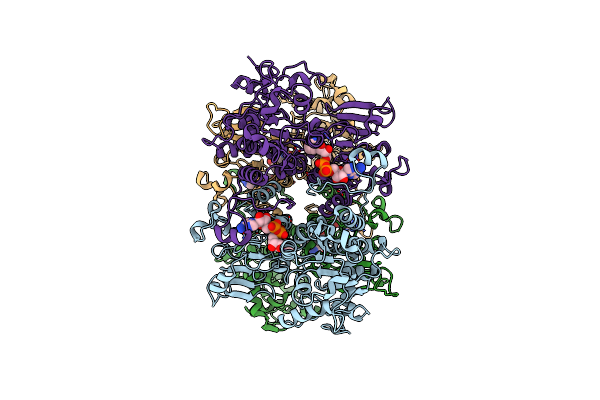 |
Cryo-Em Structure Of Pseudomonas Aeruginosa Tetrameric S-Adenosyl-L-Homocysteine Hydrolase In The Fully Open State
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: HYDROLASE Ligands: NAD |
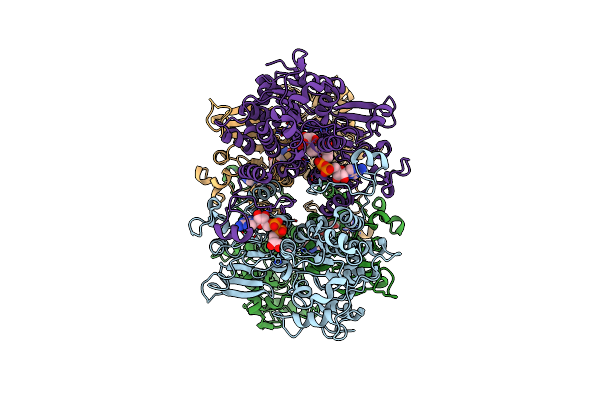 |
Cryo-Em Structure Of Pseudomonas Aeruginosa Tetrameric S-Adenosyl-L-Homocysteine Hydrolase With 3 Open And 1 Closed Subunits
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: HYDROLASE Ligands: NAD, ADN, K |
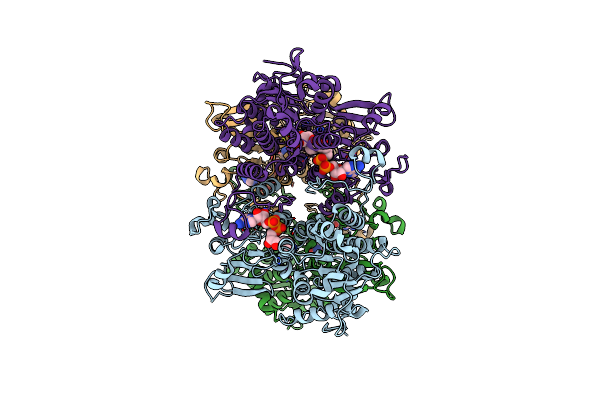 |
Cryo-Em Structure Of Pseudomonas Aeruginosa Tetrameric S-Adenosyl-L-Homocysteine Hydrolase With 1 Open And 3 Closed Subunits
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: HYDROLASE Ligands: NAD, ADN, K |
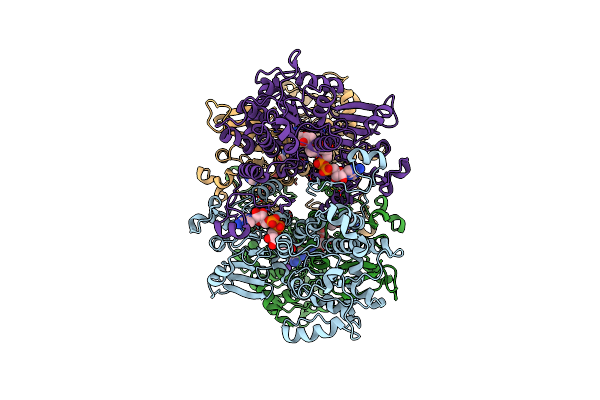 |
Cryo-Em Structure Of Pseudomonas Aeruginosa Tetrameric S-Adenosyl-L-Homocysteine Hydrolase With 2 Open And 2 Closed Subunits
Organism: Pseudomonas aeruginosa pao1
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: HYDROLASE Ligands: NAD, ADN, K |
 |
Cryo-Em Structure Of Pseudomonas Aeruginosa Tetrameric S-Adenosyl-L-Homocysteine Hydrolase With 2 Wide Open, 1 Open And 1 Closed Subunits
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: HYDROLASE Ligands: NAD, ADN |
 |
Crystal Structure Of K449E Variant Of S-Adenosyl-L-Homocysteine Hydrolase From Pseudomonas Aeruginosa In Complex With Adenosine
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2023-09-13 Classification: HYDROLASE Ligands: NAD, ADN, K, PO4 |
 |
Crystal Structure Of The F324A Mutant Of S-Adenosyl-L-Homocysteine Hydrolase From Pseudomonas Aeruginosa Cocrystallized With Adenosine In The Presence Of K+ Cations
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: NAD, ADN, K, CL, PEG |
 |
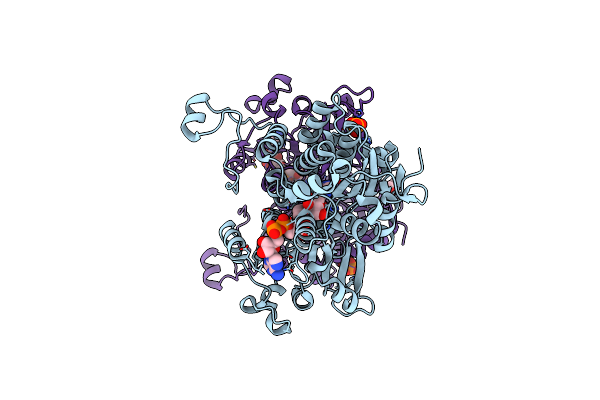
Crystal Structure Of The H323A Mutant Of S-Adenosyl-L-Homocysteine Hydrolase From Pseudomonas Aeruginosa Cocrystallized With Adenosine In The Presence Of K+ Cations
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: ADN, NAD, TRS, PEG, K, PO4, CL |
 |
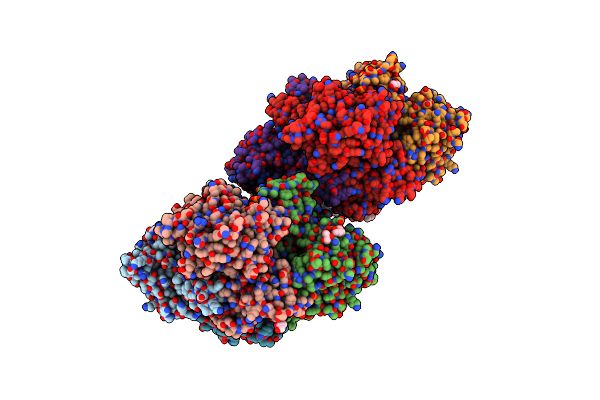
Crystal Structure Of The Q65A Mutant Of S-Adenosyl-L-Homocysteine Hydrolase From Pseudomonas Aeruginosa Cocrystallized With Adenosine In The Presence Of K+ Cations
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: PO4, NAD, ADN, GOL |
 |
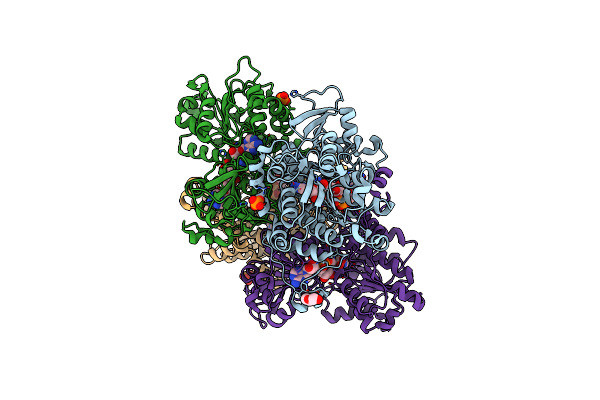
Crystal Structure Of The Q65N Mutant Of S-Adenosyl-L-Homocysteine Hydrolase From Pseudomonas Aeruginosa Crystallized In The Presence Of K+ Cations
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: NAD, GOL, PO4, K, PEG |
 |
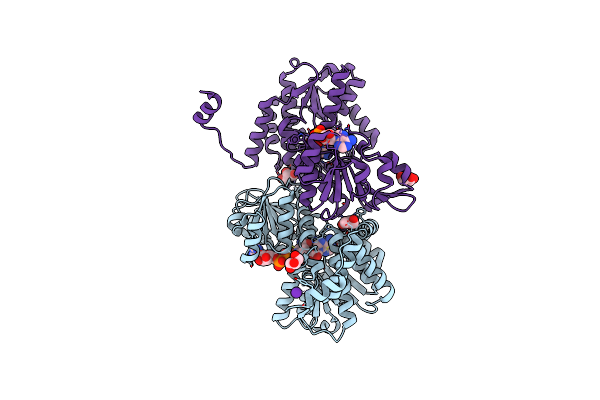
Crystal Structure Of The Q65N Mutant Of S-Adenosyl-L-Homocysteine Hydrolase From Pseudomonas Aeruginosa Cocrystallized With Adenosine In The Presence Of K+ Cations
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: NAD, ADN, PEG, PO4 |
 |
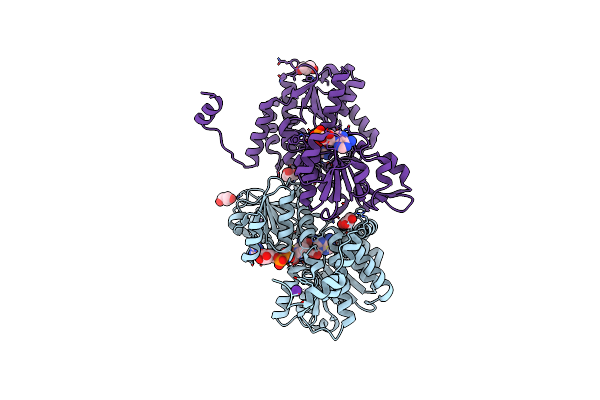
Crystal Structure Of The R24E/E352T Double Mutant Of S-Adenosyl-L-Homocysteine Hydrolase From Synechocystis Sp. Pcc 6803 Cocrystallized With Adenosine In The Presence Of Rb+ Cations
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: GOL, NAD, ADN, RB, CL |
 |
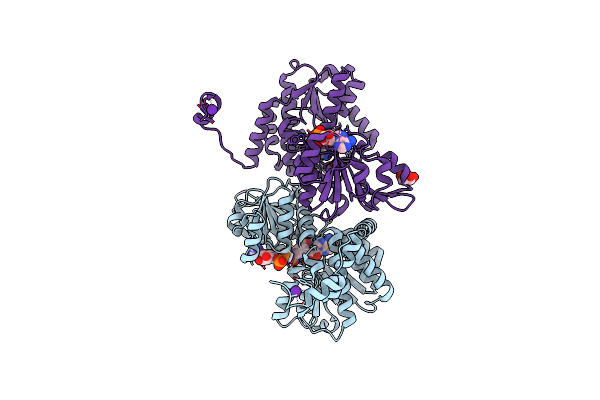
Crystal Structure Of The R24E Mutant Of S-Adenosyl-L-Homocysteine Hydrolase From Synechocystis Sp. Pcc 6803 Cocrystallized With Adenosine In The Presence Of Rb+ Cations
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: GOL, NAD, ADN, CL, RB |
 |
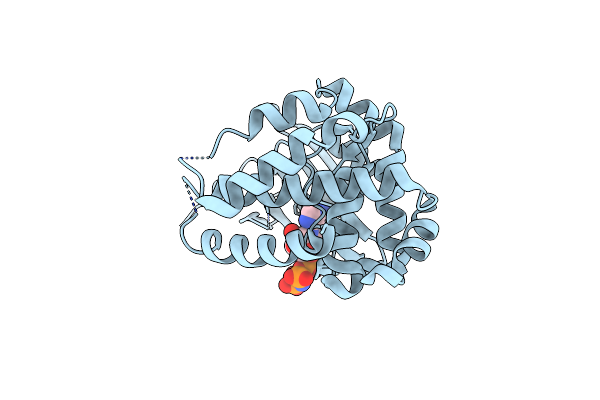
Crystal Structure Of The E352T Mutant Of S-Adenosyl-L-Homocysteine Hydrolase From Synechocystis Sp. Pcc 6803 Cocrystallized With Adenosine In The Presence Of Rb+ Cations
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-07-13 Classification: HYDROLASE Ligands: NAD, ADN, CL, RB, GOL |
 |
Organism: Bacillus subtilis py79
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-05-11 Classification: CELL CYCLE Ligands: ANP |
 |
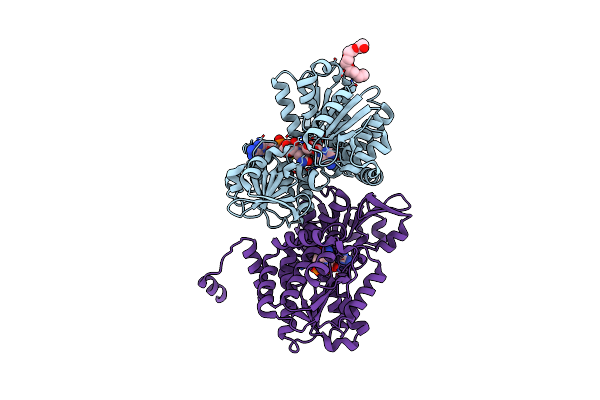
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Synechocystis Sp. Pcc 6803 Cocrystallized With Adenosine In The Presence Of Rb+ Cations
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-04-20 Classification: HYDROLASE Ligands: NAD, RB, ADN, CL |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From Synechocystis Sp. Pcc 6803 Cocrystallized With Adenosine In The Presence Of Na+ Cations
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-04-20 Classification: HYDROLASE Ligands: 1PE, NAD, ADN, CL |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadp And Malonate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: NAP, MLA, NA |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Citrate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadph And Oxalate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: OXL, NDP, GOL, CL, NA |

