Search Count: 26
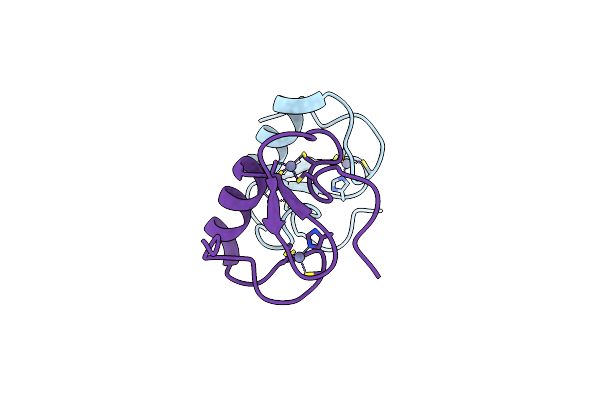 |
Crystal Structure Of The Oligomeric Form Of The Lassa Virus Matrix Protein Z
Organism: Lassa virus (strain mouse/sierra leone/josiah/1976)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-03-09 Classification: VIRAL PROTEIN Ligands: ZN |
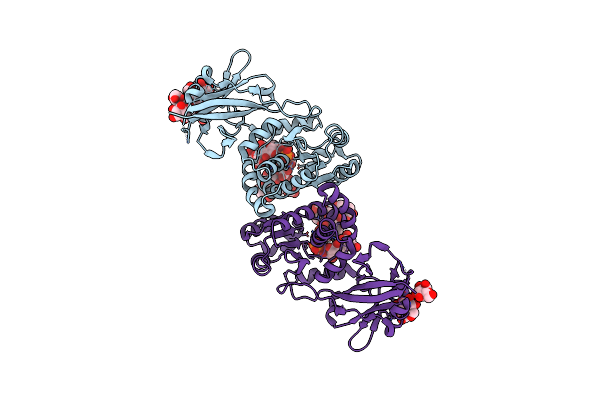 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-01-07 Classification: HYDROLASE Ligands: PO4, GLC |
 |
Organism: Toscana virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-04-09 Classification: VIRAL PROTEIN/RNA |
 |
Organism: Toscana virus
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2014-04-09 Classification: VIRAL PROTEIN |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-07-17 Classification: TRANSPORT PROTEIN Ligands: MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-05-29 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-10-03 Classification: EXOCYTOSIS Ligands: GOL |
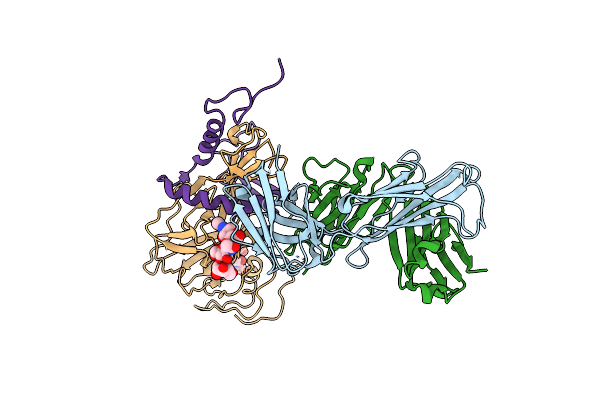 |
Crystal Structure Of Sudan Ebolavirus Glycoprotein (Strain Boniface) Bound To 16F6
Organism: Sudan ebolavirus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2012-04-18 Classification: Immune system/viral protein |
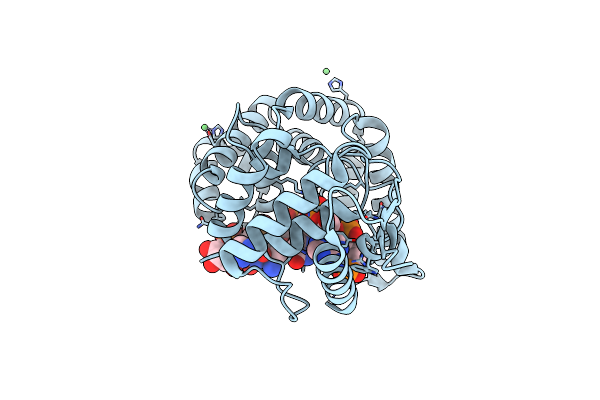 |
Organism: Mopeia lassa reassortant 29, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2012-01-11 Classification: viral protein/RNA Ligands: NI |
 |
Organism: Mopeia lassa reassortant 29, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-01-11 Classification: viral protein/RNA Ligands: PO4 |
 |
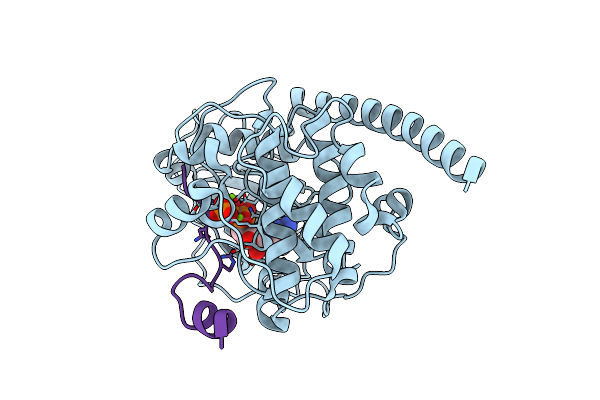
Crystal Structure Of Arg280Ala Mutant Of Catalytic Subunit Of Camp-Dependent Protein Kinase
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-12-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ATP, MG |
 |
Crystal Structure Of Glu208Ala Mutant Of Catalytic Subunit Of Camp-Dependent Protein Kinase
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2011-12-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ATP, MG |
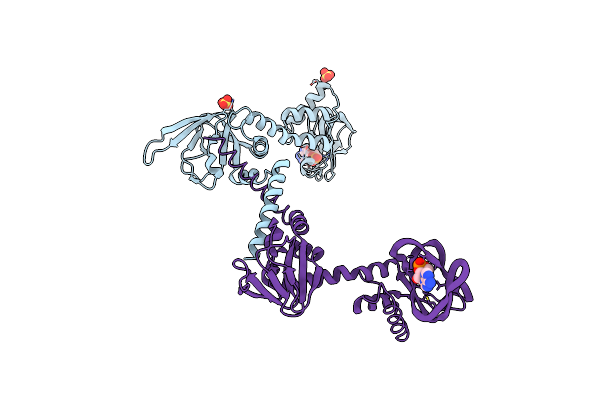 |
Crystal Structure Of Cgmp-Dependent Protein Kinase Reveals Novel Site Of Interchain Communication
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-09-21 Classification: TRANSFERASE Ligands: CMP, SO4 |
 |
Closed Crystal Structure Of Cytochrome P450 2B4 Covalently Bound To The Mechanism-Based Inactivator Tert-Butylphenylacetylene
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2011-05-11 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: HEM, TB2 |
 |
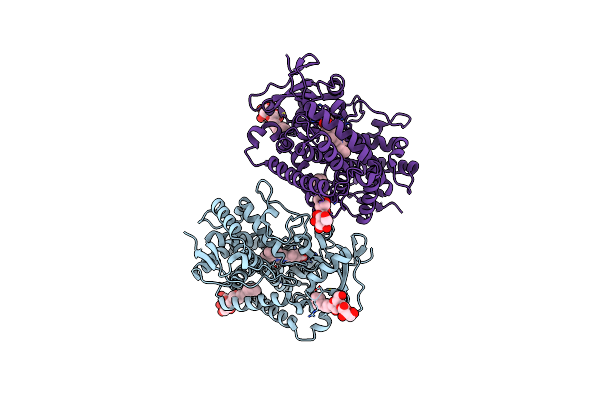
Open Crystal Structure Of Cytochrome P450 2B4 Covalently Bound To The Mechanism-Based Inactivator Tert-Butylphenylacetylene
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-05-11 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: HEM, TB2, CM5 |
 |
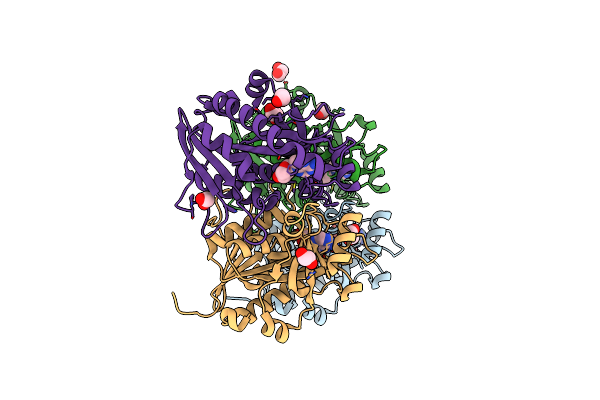
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2010-09-29 Classification: OXIDOREDUCTASE Ligands: HEM, CM5 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-06-30 Classification: HYDROLASE Ligands: SAH, ADE, EDO |
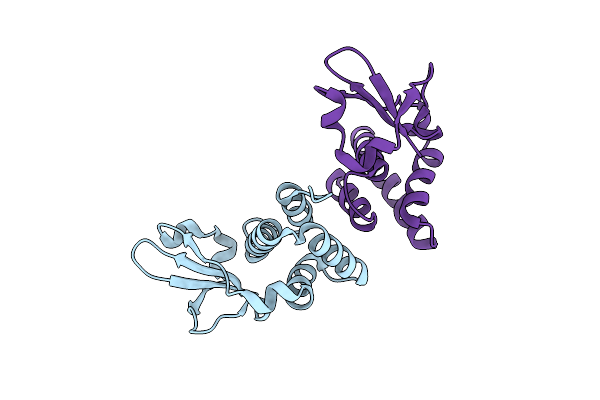 |
Organism: Reston ebolavirus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-01-12 Classification: VIRAL PROTEIN, RNA BINDING PROTEIN |
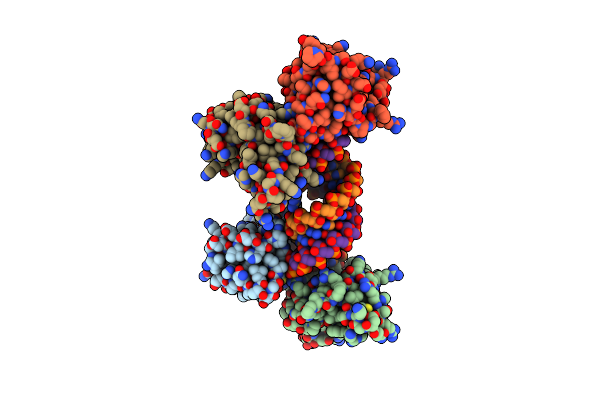 |
Crystal Structure Of Reston Ebolavirus Vp35 Rna Binding Domain In Complex With 18Bp Dsrna
Organism: Reston ebolavirus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-01-12 Classification: VIRAL PROTEIN/RNA |
 |
Crystal Structure Of A Bacteria Tir Domain, Pdtir From Paracoccus Denitrificans
Organism: Paracoccus denitrificans pd1222
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-06-16 Classification: SIGNALING PROTEIN Ligands: SO4 |

