Search Count: 14
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-04-03 Classification: CYTOKINE Ligands: XCE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-04-03 Classification: CYTOKINE Ligands: XCW, CL |
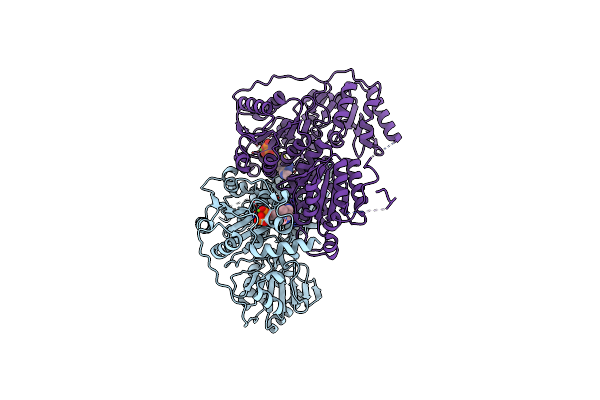 |
The Holostructure Of The Branched-Chain Keto Acid Decarboxylase (Kdca) From Lactococcus Lactis
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-12-18 Classification: LYASE Ligands: MG, TPP |
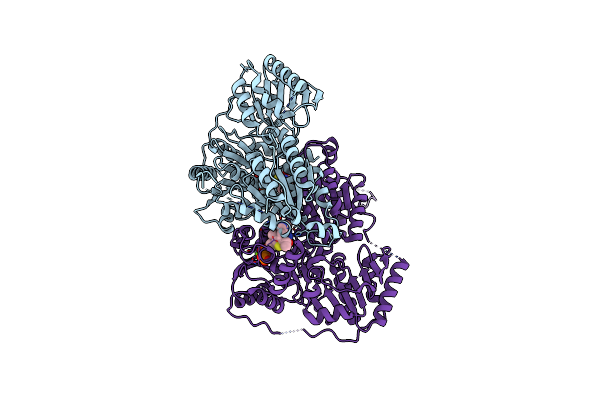 |
The Complex Structure Of The Branched-Chain Keto Acid Decarboxylase (Kdca) From Lactococcus Lactis With 2R-1-Hydroxyethyl-Deazathdp
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-11-27 Classification: LYASE Ligands: MG, R1T |
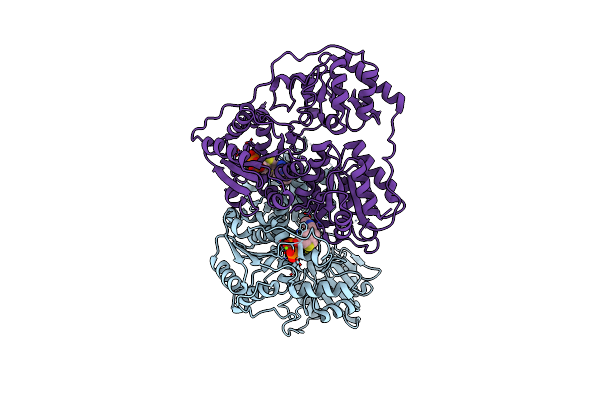 |
X-Ray Structure Of Phenylpyruvate Decarboxylase In Complex With 3-Deaza-Thdp
Organism: Azospirillum brasilense
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2007-10-23 Classification: LYASE Ligands: MG, TPW |
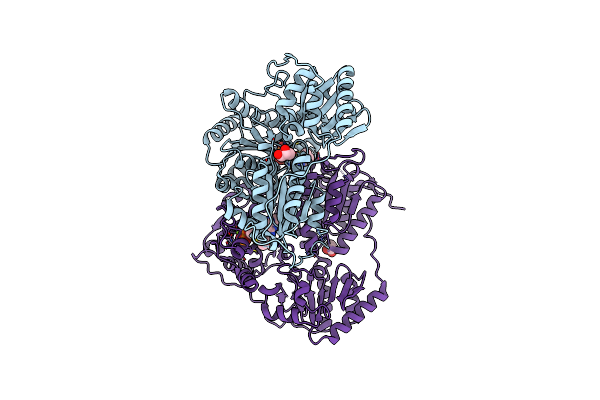 |
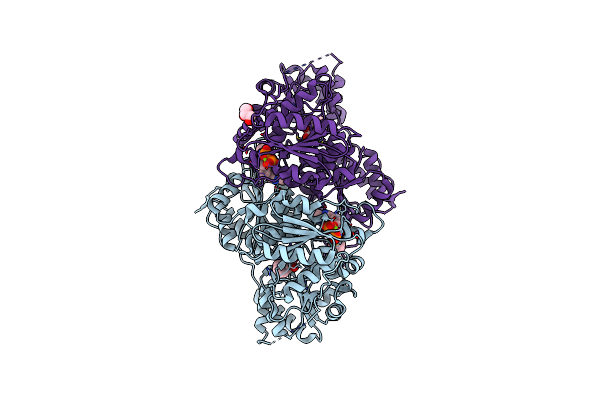
X-Ray Structure Of Phenylpyruvate Decarboxylase In Complex With 2-(1-Hydroxyethyl)-3-Deaza-Thdp
Organism: Azospirillum brasilense
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-10-23 Classification: LYASE Ligands: MG, CL, S1T, R1T, GOL |
 |
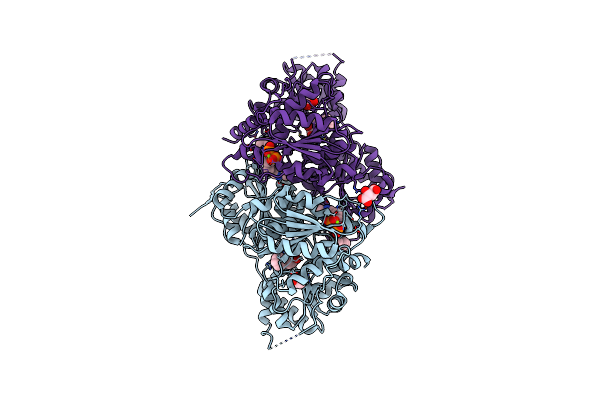
X-Ray Structure Of Phenylpyruvate Decarboxylase In Complex With 3-Deaza-Thdp And Phenylpyruvate
Organism: Azospirillum brasilense
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2007-10-23 Classification: LYASE Ligands: MG, TPW, PPY, GOL |
 |
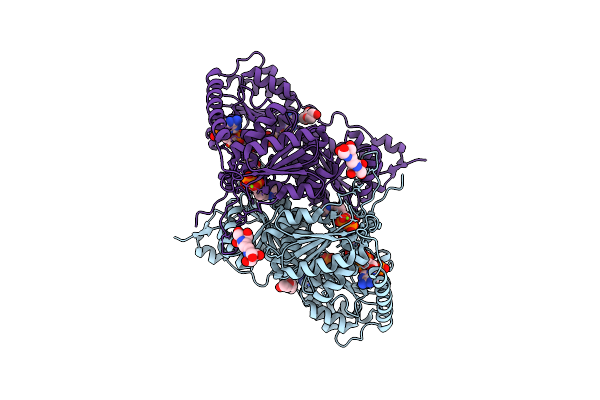
X-Ray Structure Of Phenylpyruvate Decarboxylase In Complex With 3-Deaza-Thdp And 5-Phenyl-2-Oxo-Valeric Acid
Organism: Azospirillum brasilense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-10-23 Classification: LYASE Ligands: MG, TPW, KPV, TLA, GOL |
 |
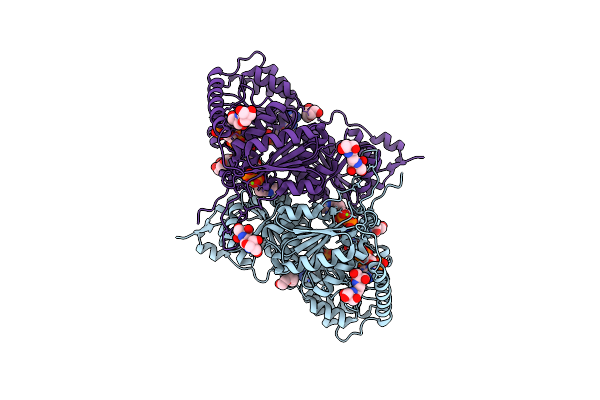
X-Ray Structure Of Oxalyl-Coa Decarboxylase In Complex With 3-Deaza- Thdp And Oxalyl-Coa
Organism: Oxalobacter formigenes
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2007-07-17 Classification: LYASE Ligands: TPW, MG, ADP, OXK, PGE, B3P |
 |
X-Ray Structure Of Oxalyl-Coa Decarboxylase With Covalent Reaction Intermediate
Organism: Oxalobacter formigenes
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2007-07-17 Classification: LYASE Ligands: OXT, MG, ADP, PGE, B3P |
 |
Organism: Oxalobacter formigenes
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2007-07-17 Classification: LYASE Ligands: TPP, MG, ADP, FYN, PGE |
 |
Organism: Oxalobacter formigenes
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-07-17 Classification: LYASE Ligands: TPW, MG, ADP, B3P |
 |
Organism: Oxalobacter formigenes
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-07-17 Classification: LYASE Ligands: TPP, MG, ADP, COA, PGE |
 |
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-03-09 Classification: Avirulence Protein |

