Search Count: 26
 |
Organism: Escherichia coli (strain k12), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-03-20 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Escherichia coli (strain k12), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-20 Classification: OXIDOREDUCTASE |
 |
Organism: Escherichia coli (strain k12), Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-03-20 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2018-11-28 Classification: TRANSFERASE Ligands: GSH, SO4, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-11-28 Classification: TRANSFERASE Ligands: SO4, ACT, MLX |
 |
Crystal Structure Of An Actin Monomer In Complex With The Nucleator Cordon-Bleu Met72Nle Wh2-Motif Peptide
Organism: Mus musculus, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-26 Classification: PEPTIDE BINDING PROTEIN Ligands: ATP, CA |
 |
Structure-Activity Studies Of Mdm2/Mdm4-Binding Stapled Peptides Comprising Non-Natural Amino Acids
Organism: Homo sapiens, Phage display vector ptdisp
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2017-12-27 Classification: ONCOPROTEIN/INHIBITOR |
 |
Organism: Aequorea victoria, Camelus dromedarius, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-12-20 Classification: FLUORESCENT PROTEIN/INHIBITOR Ligands: GOL, PO4, PG4 |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2016-02-10 Classification: TRANSFERASE Ligands: GSH |
 |
Organism: Oryza sativa subsp. indica
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2016-02-10 Classification: TRANSFERASE |
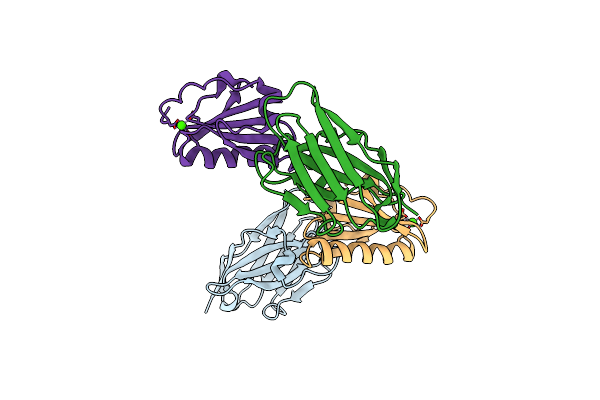 |
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2015-06-03 Classification: METAL BINDING PROTEIN/IMMUNE SYSTEM Ligands: CA |
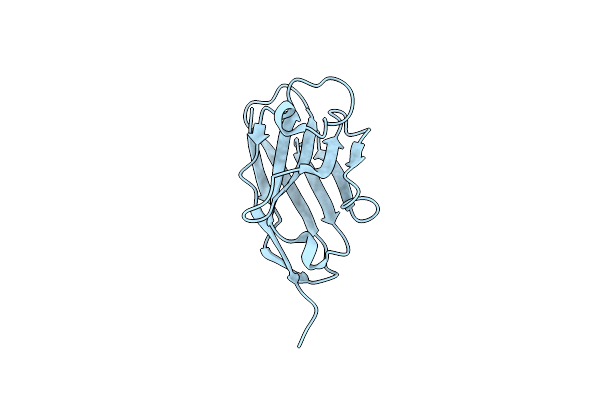 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-06-03 Classification: IMMUNE SYSTEM |
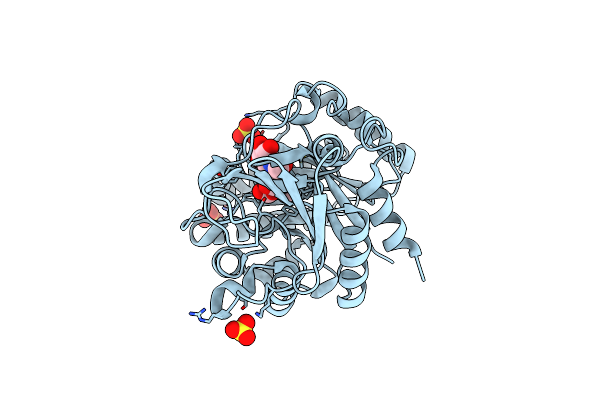 |
The Crystal Structures Of Ykl-39 In The Presence Of Chitooligosaccharides (Glcnac2) Were Solved To Resolutions Of 1.5 Angstrom
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2014-12-03 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
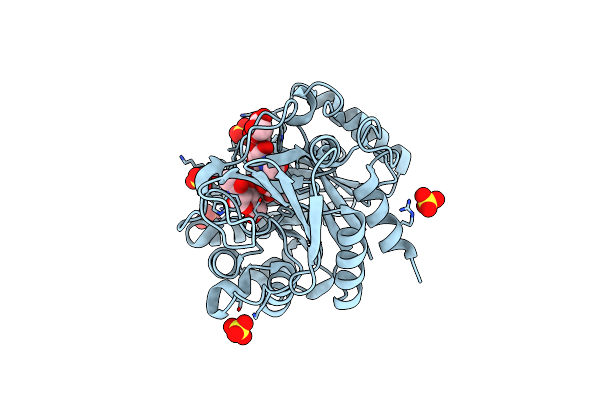 |
The Crystal Structures Of Ykl-39 In The Presence Of Chitooligosaccharides (Glcnac4) Were Solved To Resolutions Of 1.9 Angstrom
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2014-12-03 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
The Crystal Structures Of Ykl-39 In The Presence Of Chitooligosaccharides (Glcnac6) Were Solved To Resolutions Of 2.48 Angstrom
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2014-12-03 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
The Crystal Structures Of Ykl-39 In The Absence Of Chitooligosaccharides Was Solved To Resolutions Of 2.4 Angstrom
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-10-01 Classification: SUGAR BINDING PROTEIN Ligands: SO4, PEG |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-05-28 Classification: CELL CYCLE |
 |
Crystal Structure Of Glutathione Transferase Dmgstd1 From Drosophila Melanogaster, In Complex With Glutathione
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-03-30 Classification: TRANSFERASE Ligands: GSH |
 |
Crystal Structure Of Glutathione Transferase Dmgstd10 From Drosophila Melanogaster, In Complex With Glutathione
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2010-03-09 Classification: TRANSFERASE Ligands: GSH |
 |
Crystal Structure Of A Delta Class Gst (Adgstd4-4) From Anopheles Dirus, With Glutathione Complexed In One Subunit
Organism: Anopheles dirus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-02-16 Classification: TRANSFERASE Ligands: GSH |

