Search Count: 15
 |
Cryo-Em Reveals Transition States Of The Acinetobacter Baumannii F1-Atpase Rotary Subunits Gamma And Epsilon And Novel Compound Targets - Conformation 1
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: HYDROLASE Ligands: ATP, MG, ADP, PO4 |
 |
Cryo-Em Reveals Transition States Of The Acinetobacter Baumannii F1-Atpase Rotary Subunits Gamma And Epsilon And Novel Compound Targets - Conformation 2
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Cryo-Em Reveals Transition States Of The Acinetobacter Baumannii F1-Atpase Rotary Subunits Gamma And Epsilon And Novel Compound Targets - Conformation 3
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Cryo-Em Reveals Transition States Of The Acinetobacter Baumannii F1-Atpase Rotary Subunits Gamma And Epsilon And Novel Compound Targets - Conformation 4
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: HYDROLASE Ligands: ATP, MG, ADP, PO4 |
 |
Nmr Solution Structure Of Subunit Epsilon Of The Acinetobacter Baumannii F-Atp Synthase
Organism: Acinetobacter baumannii
Method: SOLUTION NMR Release Date: 2023-11-22 Classification: ELECTRON TRANSPORT |
 |
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: HYDROLASE Ligands: ATP, MG, ADP, PO4 |
 |
Cryo-Em Structure Of The Mycobacterium Tuberculosis Cytochrome Bcc:Aa3 Supercomplex And A Novel Inhibitor Targeting Subunit Cytochrome Ci
Organism: Mycobacterium tuberculosis variant bovis bcg
Method: ELECTRON MICROSCOPY Release Date: 2023-05-03 Classification: OXIDOREDUCTASE Ligands: FES, HEM, HEC, HEA |
 |
Solution Structure Of Subunit Epsilon Of The Mycobacterium Abscessus F-Atp Synthase
Organism: Mycobacteroides abscessus
Method: SOLUTION NMR Release Date: 2023-03-08 Classification: ELECTRON TRANSPORT |
 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: HYDROLASE Ligands: ATP, MG, ADP |
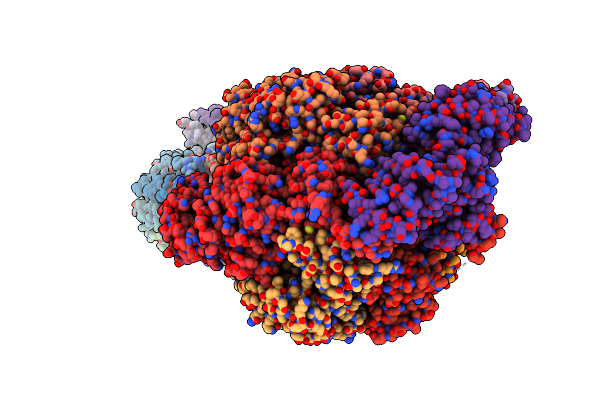 |
Cryo-Em Structure Of F-Atp Synthase From Mycolicibacterium Smegmatis (Rotational State 1)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: HYDROLASE Ligands: ATP, MG, ADP |
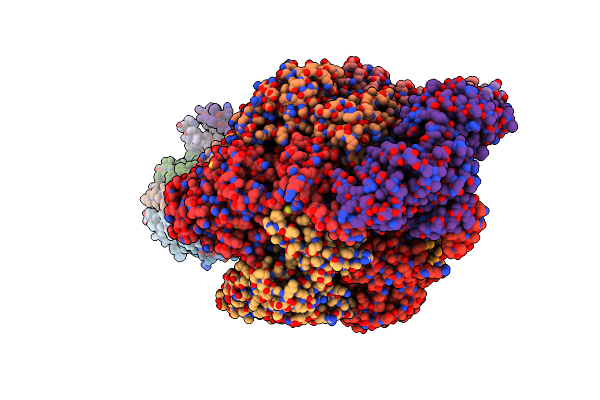 |
Cryo-Em Structure Of F-Atp Synthase From Mycolicibacterium Smegmatis (Rotational State 2)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: HYDROLASE Ligands: MG, ATP, ADP |
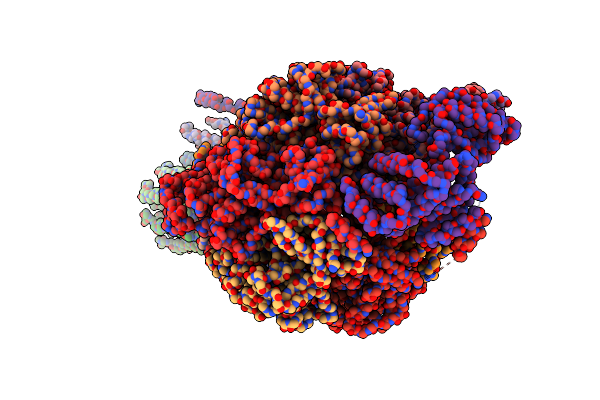 |
Cryo-Em Structure Of F-Atp Synthase From Mycolicibacterium Smegmatis (Rotational State 3) (Backbone)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: HYDROLASE |
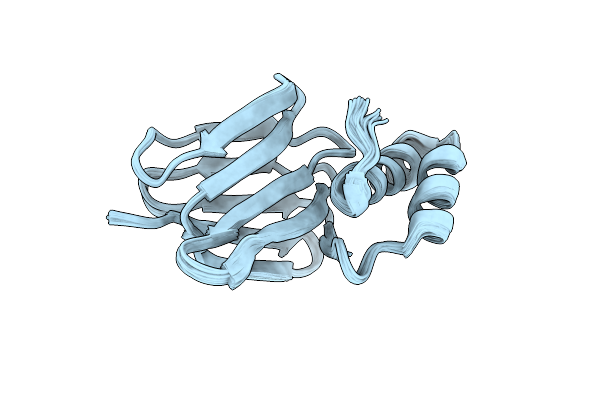 |
Solution Structure Of Subunit Epsilon Of The Mycobacterium Abscessus F-Atp Synthase
Organism: Mycobacteroides abscessus
Method: SOLUTION NMR Release Date: 2022-10-05 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: ZN, CRD, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2001-02-28 Classification: TRANSFERASE Ligands: SO4, EDO |

