Search Count: 21
 |
De Novo Designed Bbf-14 Beta Barrel With Computationally Designed Bbf-14_B4 Binder
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: DE NOVO PROTEIN Ligands: P6G, K, PG4, CL |
 |
Der F 21 Dust Mite Allergen With Computationally Designed Derf21_B10 Binder
Organism: Dermatophagoides farinae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: DE NOVO PROTEIN |
 |
Organism: Dermatophagoides farinae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: DE NOVO PROTEIN |
 |
Organism: Dermatophagoides farinae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: DE NOVO PROTEIN |
 |
Organism: Sphingobium herbicidovorans (strain atcc 700291 / dsm 11019 / nbrc 16415 / mh)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: CO, AKG |
 |
Organism: Sphingobium herbicidovorans (strain atcc 700291 / dsm 11019 / nbrc 16415 / mh)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE |
 |
Organism: Sphingobium herbicidovorans
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: FTV, CO, AKG, CL |
 |
Organism: Sphingobium herbicidovorans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: CO, AKG, CFA |
 |
Organism: Sphingobium herbicidovorans
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: CO, AKG |
 |
Organism: Sphingobium herbicidovorans
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: CO, FTJ, AKG, CL |
 |
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-07-20 Classification: TOXIN |
 |
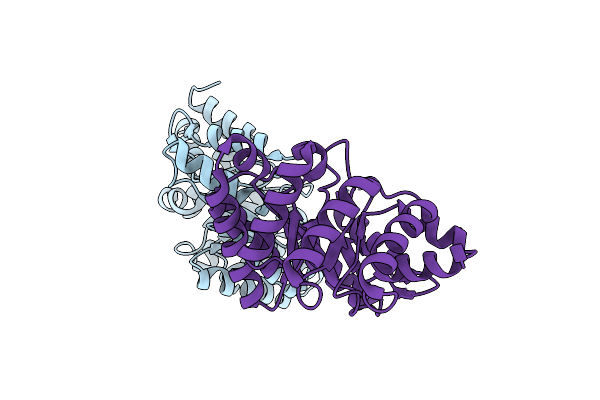
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-02-09 Classification: LYASE |
 |
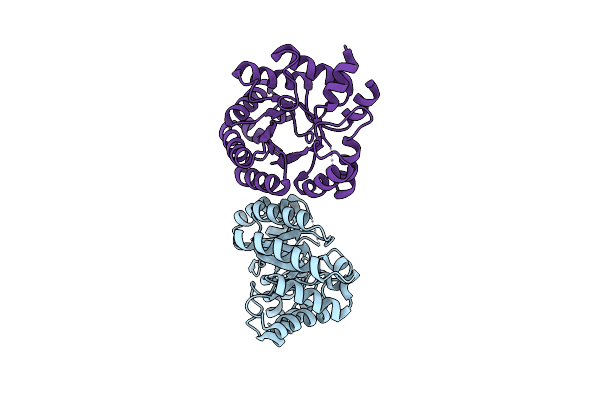
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2011-02-09 Classification: LYASE |
 |
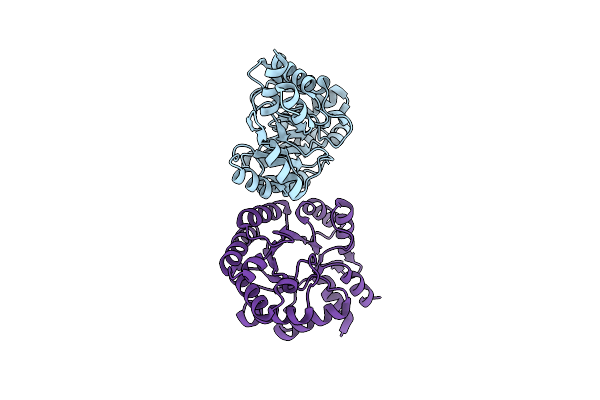
In Silico Designed Of An Improved Kemp Eliminase Ke70 Mutant By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2011-02-09 Classification: LYASE |
 |
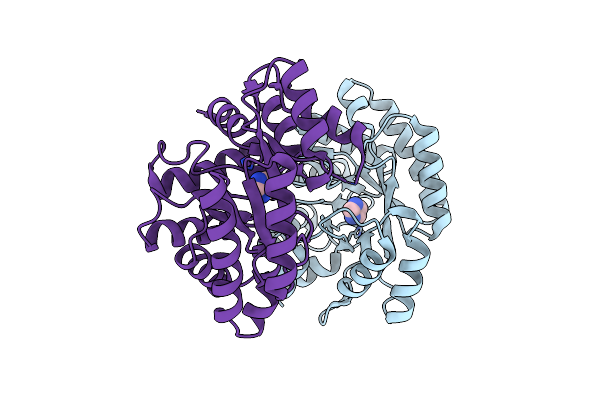
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2011-02-09 Classification: LYASE |
 |
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution R2 3/5G
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2011-02-09 Classification: LYASE Ligands: IMD |
 |
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution R4 8/5A
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2011-02-09 Classification: LYASE Ligands: NO3, BEN |
 |
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution R5 7/4A
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-02-09 Classification: LYASE Ligands: BEN |
 |
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution R6 6/10A
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2011-02-09 Classification: LYASE |
 |
Optimization Of The In Silico Designed Kemp Eliminase Ke70 By Computational Design And Directed Evolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2011-02-09 Classification: LYASE Ligands: 3NY |

