Search Count: 26
 |
Organism: Listeria monocytogenes 10403s
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: TOXIN |
 |
Arsenate Reductase (Arsc2) From Deinococcus Indicus, Co-Crystallized With Arsenate
Organism: Deinococcus indicus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE Ligands: ART, GOL |
 |
Organism: Deinococcus indicus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-08-30 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of The Albicidin Resistance Protein Stm3175 From Salmonella Typhimurium
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2022-09-28 Classification: DNA BINDING PROTEIN |
 |
Estimation Of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2019-01-30 Classification: CHAPERONE Ligands: D4W |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2017-08-23 Classification: CHAPERONE PROTEIN Ligands: 73S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-03-23 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2013-11-13 Classification: LIGASE/LIGASE INHIBITOR Ligands: Y30, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2013-11-13 Classification: LIGASE/LIGASE INHIBITOR Ligands: 28W |
 |
Variable Internal Flexibility Characterizes The Helical Capsid Formed By Agrobacterium Vire2 Protein On Single-Stranded Dna.
Organism: Agrobacterium tumefaciens
Method: ELECTRON MICROSCOPY Resolution:20.00 Å Release Date: 2013-06-26 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-09-12 Classification: PROTEIN BINDING/INHIBITOR Ligands: TJ2, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-11-02 Classification: LIGASE Ligands: 07G |
 |
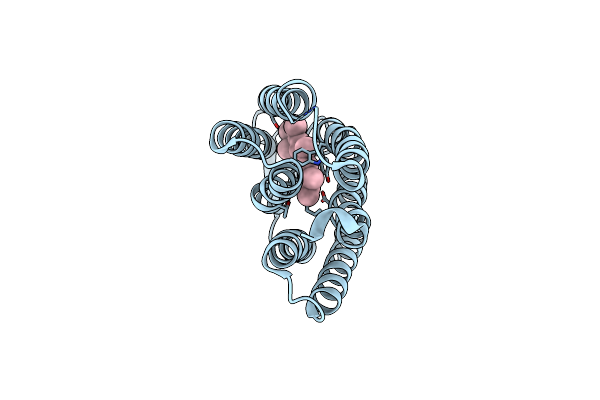
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-08-25 Classification: PROTON TRANSPORT Ligands: RET, LI1 |
 |
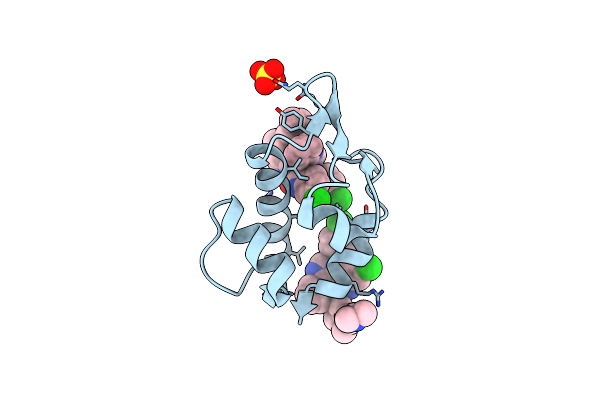
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2010-08-25 Classification: PROTON TRANSPORT Ligands: RET |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-03-16 Classification: CELL CYCLE Ligands: WW8, SO4 |
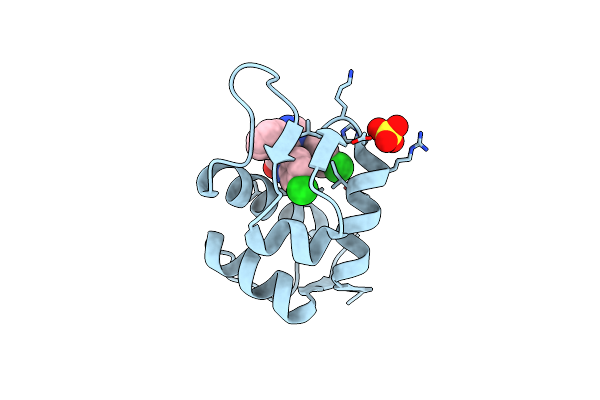 |
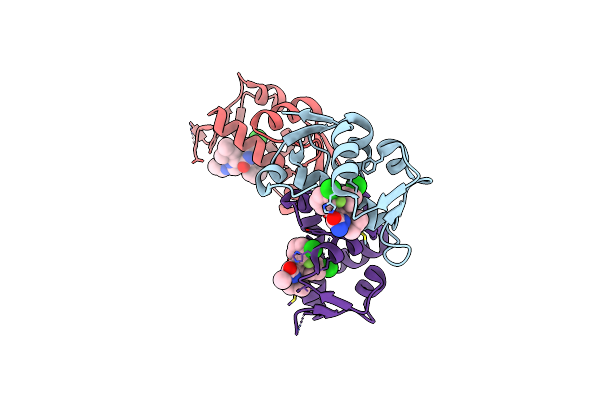
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-03-16 Classification: LIGASE Ligands: K23, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-03-16 Classification: LIGASE Ligands: MI6 |
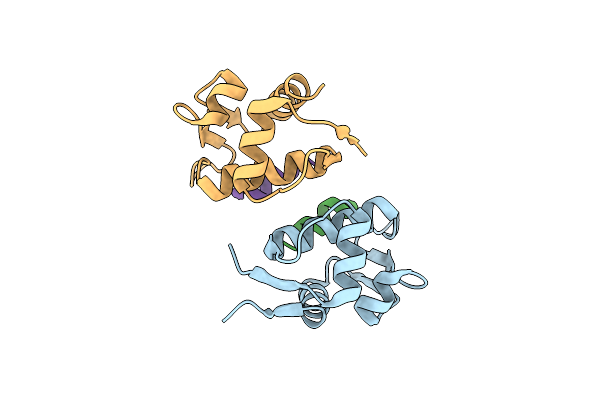 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-04-14 Classification: CELL CYCLE |
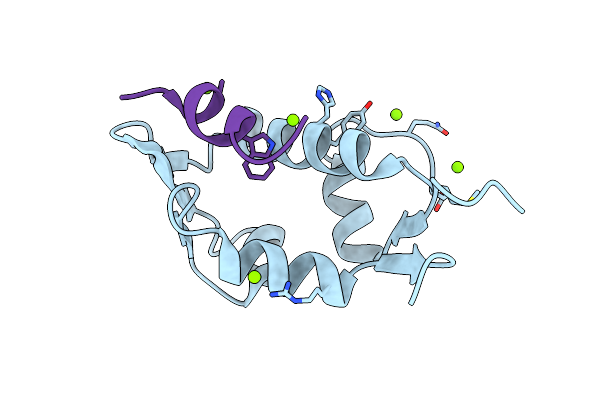 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-12-16 Classification: CELL CYCLE Ligands: MG |
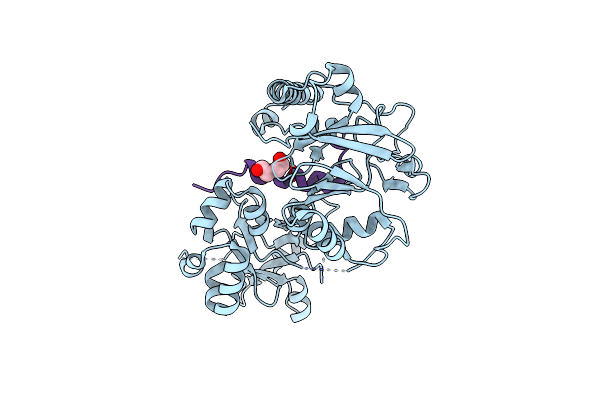 |
Crystal Structure Of Agrobacterium Tumefaciens Vire2 In Complex With Its Chaperone Vire1: A Novel Fold And Implications For Dna Binding
Organism: Agrobacterium tumefaciens str.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-08-19 Classification: DNA BINDING PROTEIN, CHAPERONE Ligands: NH4, PEG |

