Search Count: 426
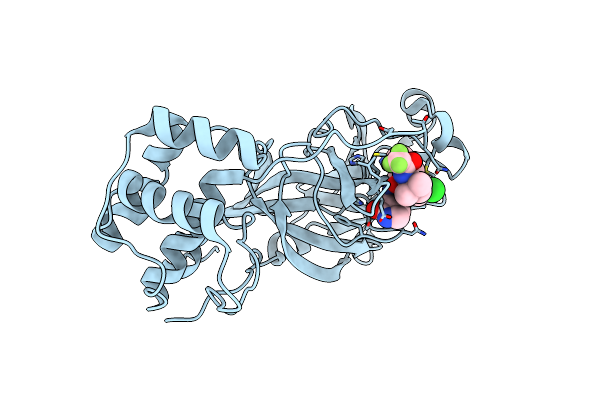 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-277-5Cl
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: A1B7C |
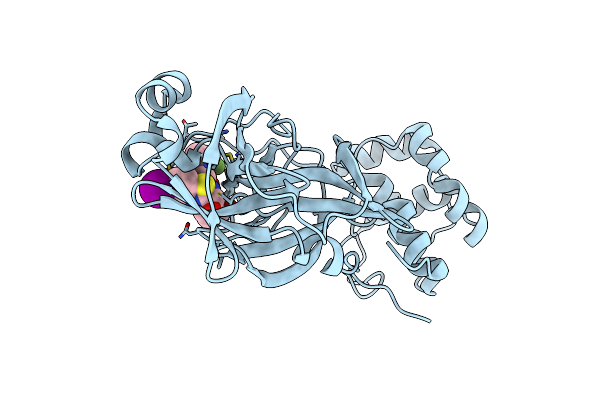 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-280-5I
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: A1B7A |
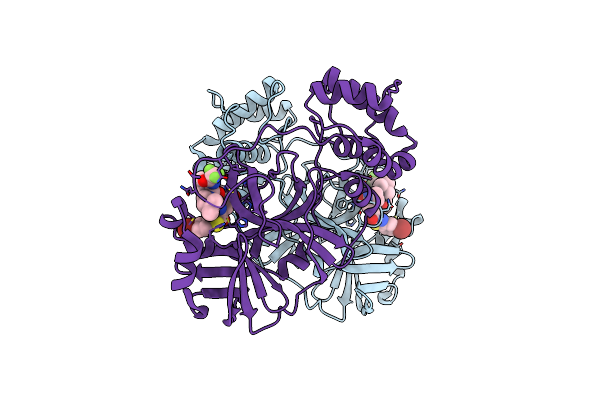 |
Crystal Structure Of Sars-Cov-2 Main Protease In Complex With An Inhibitor Tkb-276-5Br
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: A1B7B |
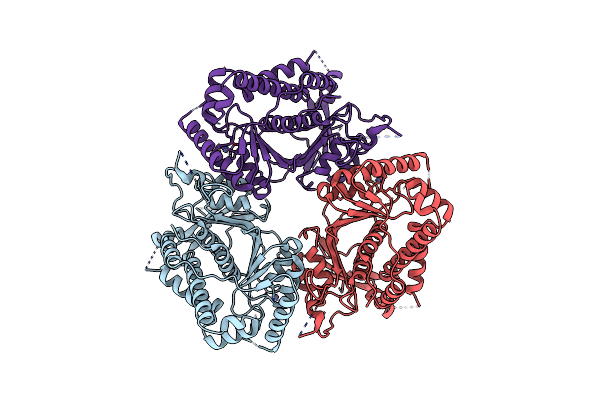 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Trimer Without A Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN |
 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer Without A Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN |
 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer With Dctp Bound In The Active Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN, DCP |
 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer With 5Mdctp Bound In The Active Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN, A1I2I |
 |
Organism: Thermococcus kodakarensis
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: ISOMERASE |
 |
Crystallization And Structural Characterization Of Phosphopentomutase From The Hyperthermophilic Archaeon Thermococcus Kodakarensis
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2024-12-18 Classification: ISOMERASE Ligands: 7PE, GOL |
 |
The Crystal Structure Of A Human Monoclonal Antibody (Aab), Termed Tg10, Used To Study Poly-N-Acetyl-Glucosamine Broadly Expressed In Biofilm-Forming Pathogenclonal Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-12-04 Classification: IMMUNE SYSTEM Ligands: SO4, NA |
 |
The Crystal Structure Of A Human Monoclonal Antibody (Aab), Termed Tg10, Complexed With A Glcnh2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-12-04 Classification: IMMUNE SYSTEM Ligands: SO4, CL, GCS |
 |
The Crystal Structure Of A Human Monoclonal Antibody (Aab), Termed Tg10, Complexed With A Disaccharide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-12-04 Classification: IMMUNE SYSTEM Ligands: SO4, CL |
 |
Structure Of A Type Iii Antifreeze Protein Isoform Hplc12 Re-Refined Using Standard Protocols
Organism: Zoarces americanus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-07-17 Classification: ANTIFREEZE PROTEIN |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Authentic Protein) In Complex With An Inhibitor Tkb-245
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: T2L |
 |
Crystal Structure Of Sars-Cov-2 Main Protease E166V Mutant In Complex With An Inhibitor Tkb-245
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: T2L |
 |
Crystal Structure Of Sars-Cov-2 Main Protease A191T Mutant In Complex With An Inhibitor 5H
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: V7G |
 |
Crystal Structure Of Sars-Cov-2 Main Protease A191T Mutant In Complex With An Inhibitor Nirmatrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-04-24 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI, PEG |
 |
Organism: Rhodospirillum rubrum atcc 11170
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: CL, GOL |
 |
Structure Of Atypical Asparaginase From Rhodospirillum Rubrum (Mutant K19E)
Organism: Rhodospirillum rubrum atcc 11170
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: CL, EDO |
 |
Structure Of Atypical Asparaginase From Rhodospirillum Rubrum In Complex With L-Asp
Organism: Rhodospirillum rubrum atcc 11170
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: ASP, CL |

