Search Count: 35
 |
Molecular Basis Of Zp3/Zp1 Heteropolymerization: Crystal Structure Of A Native Vertebrate Egg Coat Filament
Organism: Oryzias latipes
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-03-13 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of The Complete N-Terminal Region Of Human Zp2 (Hzp2-N1N2N3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-03-13 Classification: CELL ADHESION Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-03-13 Classification: CELL ADHESION Ligands: NAG |
 |
Crystal Structure Of Tetrameric Collagenase-Cleaved Xenopus Zp2-N2N3 (Cleaved Xzp2-N2N3)
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-03-13 Classification: CELL ADHESION Ligands: NAG, NO3, BCN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-03-13 Classification: CELL ADHESION Ligands: NAG |
 |
Molecular Basis Of Zp3/Zp1 Heteropolymerization: Crystal Structure Of A Native Vertebrate Egg Coat Filament Fragment
Organism: Oryzias latipes
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2024-03-13 Classification: STRUCTURAL PROTEIN Ligands: YB |
 |
Full-Length Bacterial Polysaccharide Co-Polymerase Wzze From E. Coli. C4 Symmetry
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN |
 |
Full-Length Bacterial Polysaccharide Co-Polymerase Wzze Mutant R267A From E. Coli. C4 Symmetry
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN |
 |
Full-Length Bacterial Polysaccharide Co-Polymerase Wzze Mutant R267E From E. Coli. C4 Symmetry
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: MEMBRANE PROTEIN |
 |
Cryoem Structure Of The Abc Transporter Bmra E504A Mutant In Complex With Atp-Mg
Organism: Bacillus subtilis (strain 168)
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
 |
Multidrug Resistance Transporter Bmra Mutant E504A Bound With Atp, Mg, And Rhodamine 6G Solved By Cryo-Em
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, RHQ |
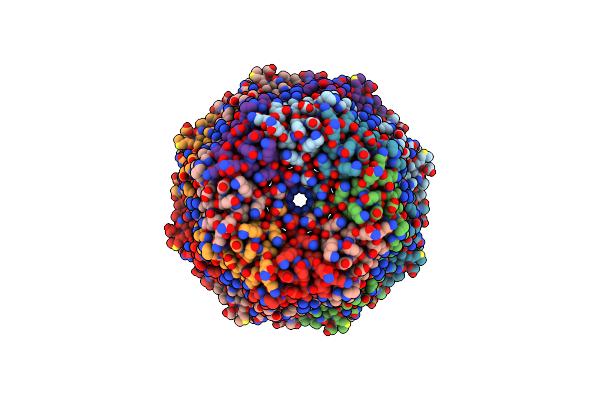 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: MEMBRANE PROTEIN |
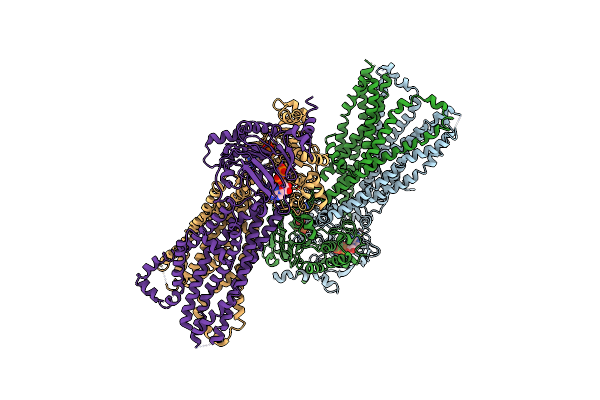 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.95 Å Release Date: 2020-05-06 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
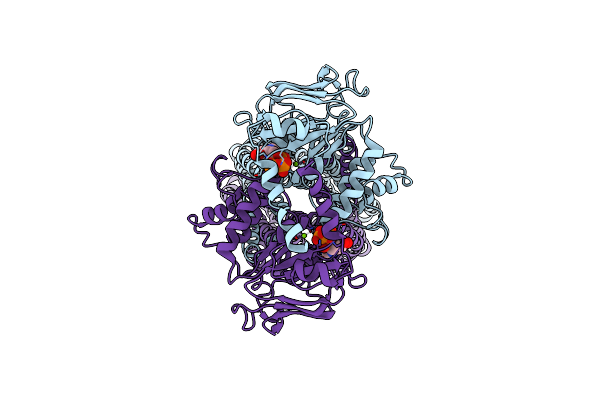 |
Multidrug Resistance Transporter Bmra Mutant E504A Bound With Atp And Mg Solved By Cryo-Em
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2020-05-06 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
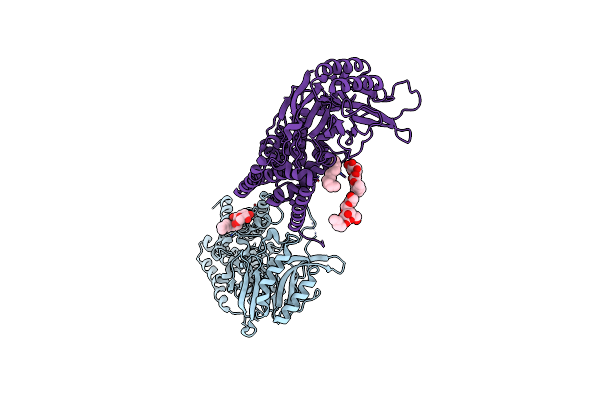 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-01-29 Classification: MEMBRANE PROTEIN Ligands: LMT, D12 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2019-12-18 Classification: MEMBRANE PROTEIN Ligands: GOL, OLC |
 |
Structure Of A Functional Obligate Respiratory Supercomplex From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis mc2 155, Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: ELECTRON MICROSCOPY Release Date: 2018-11-07 Classification: ELECTRON TRANSPORT Ligands: FES, CDL, MQ9, CU, HAS, HEC, HEM |
 |
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-07 Classification: OXIDOREDUCTASE Ligands: HEM, NA, OXY, MPD, PO4 |
 |
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-09-07 Classification: OXIDOREDUCTASE Ligands: HEM, NA, OXY, MPD, TRS, PO4 |
 |
Organism: Burkholderia pseudomallei (strain 1710b)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2016-09-07 Classification: OXIDOREDUCTASE Ligands: HEM, NA, CL, OXY, MPD, MRD, ADP |

