Search Count: 28
 |
Cryo-Em Structure Of Rat Cardiac Sodium Channel Nav1.5 With Batrachotoxin Analog Btx-B
Organism: Rattus norvegicus, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: MEMBRANE PROTEIN Ligands: NAG, LBN, Y01, 9Z9, YIJ |
 |
Crystal Structure Of Navab I22V As A Basis For The Human Nav1.7 Inherited Erythromelalgia I136V Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: BGC, PX4, CPS |
 |
Crystal Structure Of Navab E96P As A Basis For The Human Nav1.7 Inherited Erythromelalgia S211P Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: PX4, CPS |
 |
Structure Of Navab L98R As A Basis For The Human Nav1.7 Inherited Erythromelalgia L823R Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: PX4 |
 |
Crystal Structure Of Navab L101S As A Basis For The Human Nav1.7 Inherited Erythromelalgia F216S Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: PX4, CPS |
 |
Crystal Structure Of Navab I119T As A Basis For The Human Nav1.7 Inherited Erythromelalgia I234T Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-04-12 Classification: MEMBRANE PROTEIN Ligands: PX4 |
 |
Crystal Structure Of Navab L123T As A Basis For The Human Nav1.7 Inherited Erythromelalgia I848T Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-04-12 Classification: MEMBRANE PROTEIN Ligands: PX4, CPS |
 |
Crystal Structure Of Navab V126T As A Basis For The Human Nav1.7 Inherited Erythromelalgia S241T Mutation
Organism: Aliarcobacter butzleri rm4018
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-04-12 Classification: MEMBRANE PROTEIN Ligands: PX4, CPS |
 |
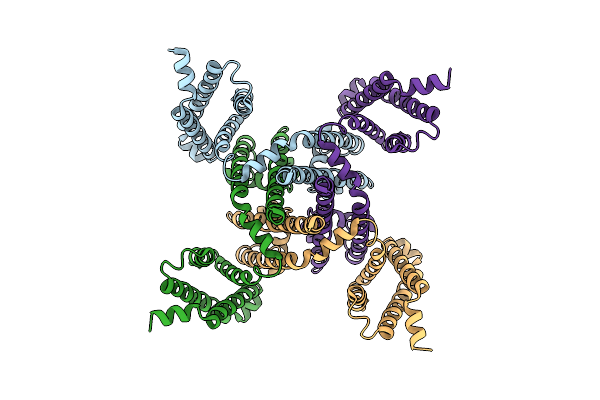
Structure Of Navab/Nav1.7-Vs2A Chimera Trapped In The Resting State By Tarantula Toxin M3-Huwentoxin-Iv
Organism: Escherichia coli (strain k12), Arcobacter butzleri (strain rm4018), Homo sapiens, Haplopelma schmidti
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: MEMBRANE PROTEIN |
 |
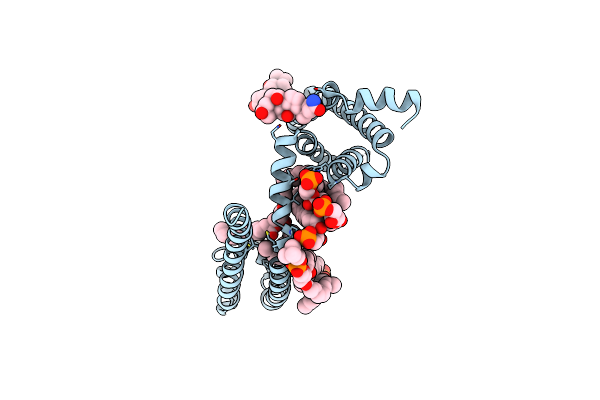
Cryo-Em Structure Of Voltage-Gated Sodium Channel Navab N49K/L109A/M116V/G94C/Q150C Disulfide Crosslinked Mutant In The Resting State
Organism: Escherichia coli k-12, Arcobacter butzleri (strain rm4018)
Method: ELECTRON MICROSCOPY Release Date: 2019-08-14 Classification: membrane protein, metal transport |
 |
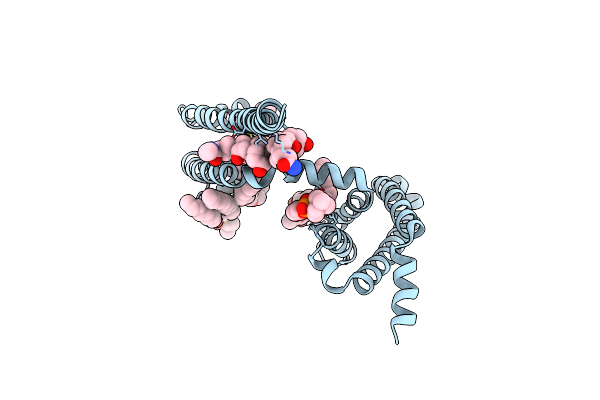
Crystal Structure Of Voltage-Gated Sodium Channel Navab G94C/Q150C Mutant In The Activated State
Organism: Arcobacter butzleri (strain rm4018)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2019-08-14 Classification: membrane protein, metal transport Ligands: PX4, CPS |
 |
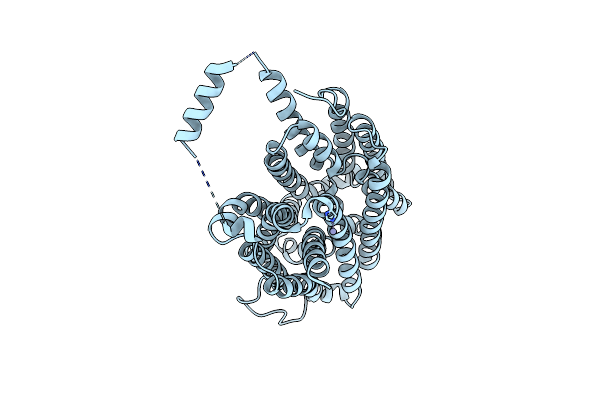
Crystal Structure Of Voltage-Gated Sodium Channel Navab V100C/Q150C Disulfide Crosslinked Mutant In The Activated State
Organism: Arcobacter butzleri (strain rm4018)
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2019-08-14 Classification: membrane protein, metal transport Ligands: CPS, PX4 |
 |
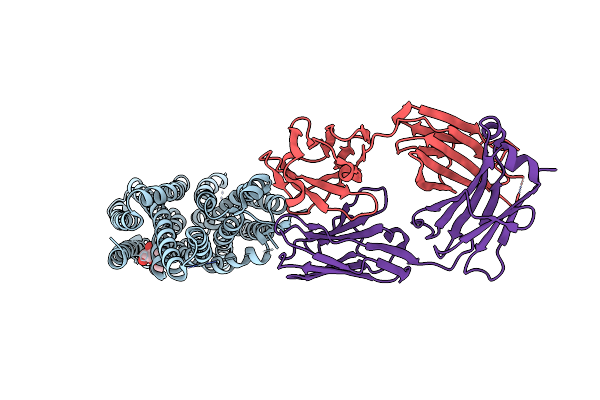
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2014-08-06 Classification: TRANSPORT PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-05-15 Classification: TRANSPORT PROTEIN/Immune System Ligands: GYP |
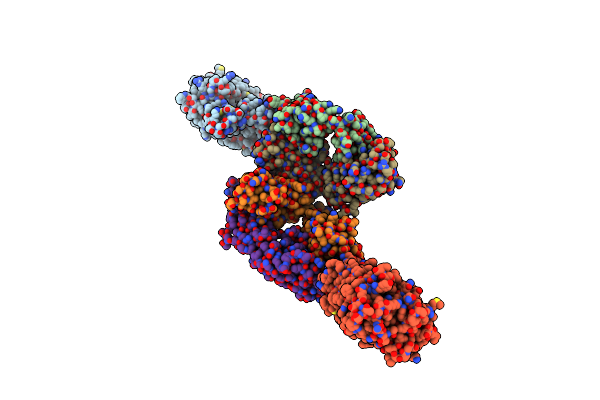 |
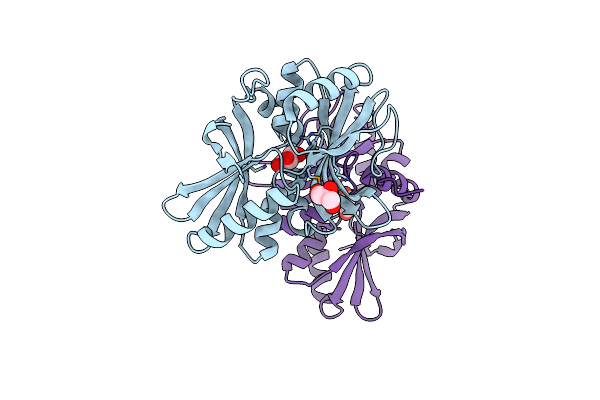
Organism: Escherichia coli, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-05-15 Classification: TRANSPORT PROTEIN/Immune System Ligands: NO2, GYP |
 |
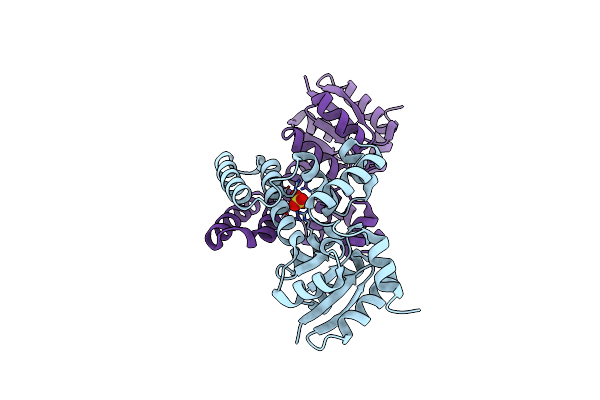
Crystal Structure Of A Nitrogen Repressor-Like Protein Mj0159 From Methanococcus Jannaschii
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-06-23 Classification: structural genomics, unknown function Ligands: GOL |
 |
Crystal Structure Of The Mycobacterium Tuberculosis Hypoxic Response Regulator Dosr
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-04-22 Classification: TRANSCRIPTION Ligands: SO4 |
 |
Crystal Structure Of The Mycobacterium Tuberculosis Hypoxic Response Regulator Dosr C-Terminal Domain Crystal Form Ii
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-04-22 Classification: TRANSCRIPTION |
 |
Crystal Structure Of The Nickel-Activated Two-Domain Iron-Dependent Regulator (Ider)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2007-02-13 Classification: TRANSCRIPTION Ligands: NI, PO4 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-02-13 Classification: TRANSCRIPTION/DNA Ligands: NI, NA |

