Search Count: 45
 |
Organism: Deinococcus radiodurans r1
Method: ELECTRON MICROSCOPY Release Date: 2022-07-13 Classification: STRUCTURAL PROTEIN |
 |
Organism: Deinococcus radiodurans r1
Method: ELECTRON MICROSCOPY Release Date: 2022-07-13 Classification: STRUCTURAL PROTEIN Ligands: JPI, JQ6, JPX, CU, FE |
 |
Native Structure Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Structure Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals Soaked With Dtt At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Structure Of Mosquitocidal Cyt1Aa Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 10
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-10-14 Classification: TOXIN Ligands: CA |
 |
Structure Of The C7S Mutant Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2019-12-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2019-12-04 Classification: TRANSPORT PROTEIN Ligands: NAI |
 |
Crystal Structure Of The Outer Membrane Channel Dcap Of Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-11-14 Classification: MEMBRANE PROTEIN |
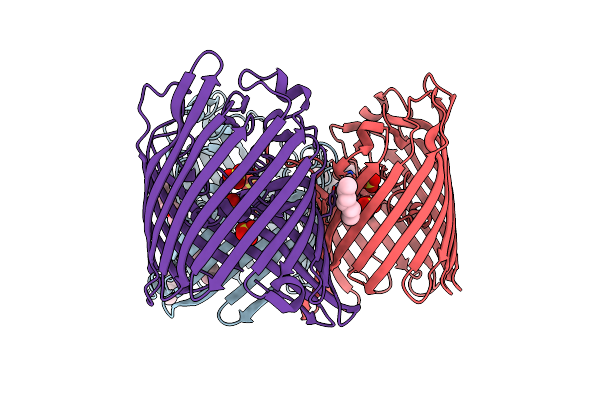 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-10-31 Classification: TRANSPORT PROTEIN Ligands: SO4, BOG, C8E |
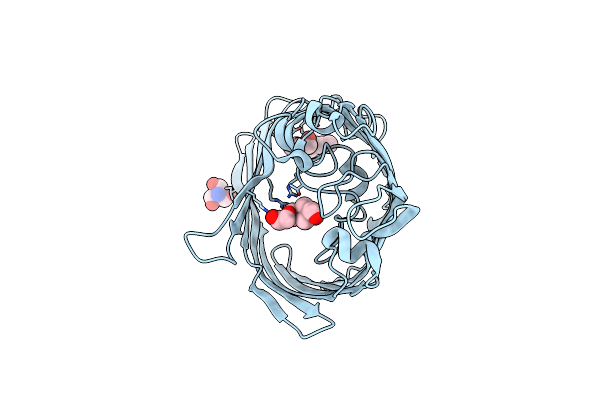 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-06-20 Classification: MEMBRANE PROTEIN Ligands: TRS, C8E |
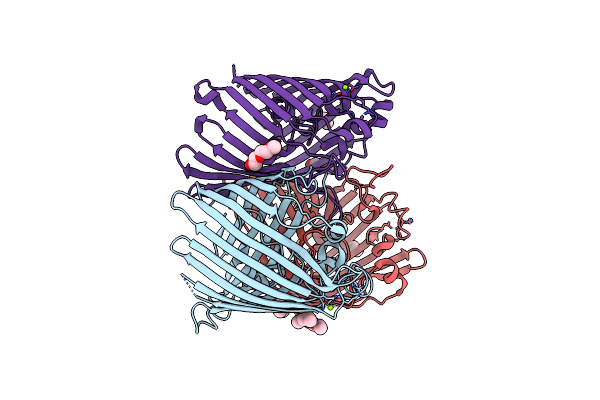 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-06-20 Classification: MEMBRANE PROTEIN Ligands: C8E, MG |
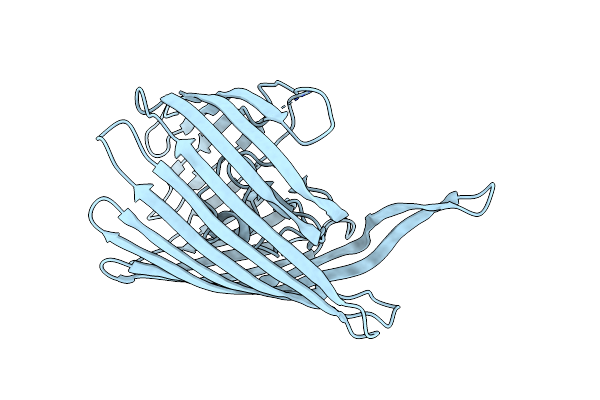 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-04-25 Classification: MEMBRANE PROTEIN |
 |
Organism: Vibrio cholerae serotype o1 (strain atcc 39541 / classical ogawa 395 / o395)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-04-25 Classification: MEMBRANE PROTEIN Ligands: C8E |
 |
Ompudeltan (N-Terminus Deletion Mutant Of Ompu), Outer Membrane Protein Of Vibrio Cholerae
Organism: Vibrio cholerae serotype o1 (strain atcc 39541 / classical ogawa 395 / o395)
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2018-04-25 Classification: MEMBRANE PROTEIN Ligands: C8E |
 |
Organism: Vibrio cholerae serotype o1 (strain atcc 39541 / classical ogawa 395 / o395)
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2018-04-25 Classification: MEMBRANE PROTEIN Ligands: C8E, MES |
 |
Omptdeltal8 (Loop L8 Deletion Mutant Of Ompt), An Outer Membrane Protein Of Vibrio Cholerae
Organism: Vibrio cholerae serotype o1 (strain atcc 39541 / classical ogawa 395 / o395)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2018-04-25 Classification: MEMBRANE PROTEIN Ligands: C8E, MG, ACT |
 |
Ompt (In-Vitro Folded), An Outer Membrane Protein Vibrio Cholerae (Trimer Form)
Organism: Vibrio cholerae serotype o1 (strain atcc 39541 / classical ogawa 395 / o395)
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2018-04-25 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Structure Of 283-Lgny-286, The Steric Zipper That Supports The Self-Association Of P. Stuartii Omp-Pst2 Into Dimers Of Trimers
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2018-02-21 Classification: CELL ADHESION Ligands: SO4 |
 |
Structure Of 206-Gvvtse-211, The Steric Zipper That Supports The Self-Association Of P. Stuartii Omp-Pst1 Into Dimers Of Trimers
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2018-02-21 Classification: CELL ADHESION |

