Search Count: 14
 |
Crystal Structure Of Extracellular Domain 1 (Ecd1) Of Ftsx From S. Pneumonie In Complex With Dodecane-Trimethylamine
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-04-24 Classification: CELL CYCLE Ligands: CAT |
 |
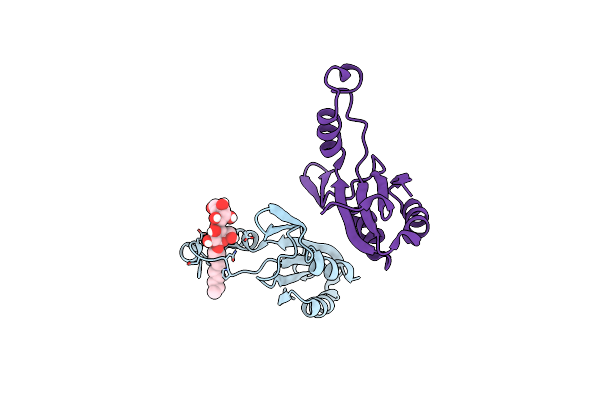
Crystal Structure Of Extracellular Domain 1 (Ecd1) Of Ftsx From S. Pneumonie In Complex With Undecyl-Maltoside
Organism: Streptococcus pneumoniae serotype 2 (strain d39 / nctc 7466)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-04-24 Classification: CELL CYCLE Ligands: UMQ, SO4, TRS |
 |
Crystal Structure Of Extracellular Domain 1 (Ecd1) Of Ftsx From S. Pneumonie In Complex With N-Decyl-B-D-Maltoside
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: DMU |
 |
Solution Structure Of The Large Extracellular Loop Of Ftsx In Streptococcus Pneumoniae
Organism: Streptococcus pneumoniae serotype 2 (strain d39 / nctc 7466)
Method: SOLUTION NMR Release Date: 2019-02-13 Classification: MEMBRANE PROTEIN |
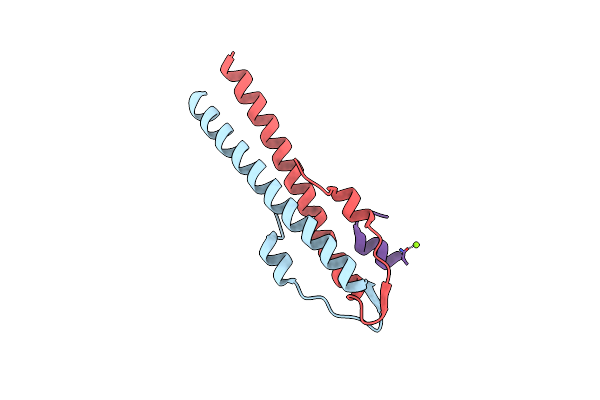 |
Cell Division Regulator, B. Subtilis Gpsb, In Complex With Peptide Fragment Of Penicillin Binding Protein Pbp1A
Organism: bacillus subtilis subsp. subtilis str. 168, Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-01-23 Classification: CELL CYCLE Ligands: MG |
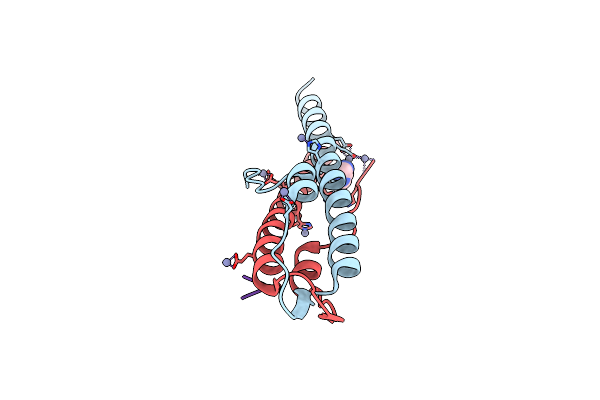 |
Cell Division Regulator Gpsb In Complex With Peptide Fragment Of L. Monocytogenes Penicillin Binding Protein Pbpa1
Organism: bacillus subtilis subsp. subtilis str. 168, Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-01-23 Classification: CELL CYCLE Ligands: ZN, IMD |
 |
Organism: Streptococcus pneumoniae r6
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-01-23 Classification: CELL CYCLE |
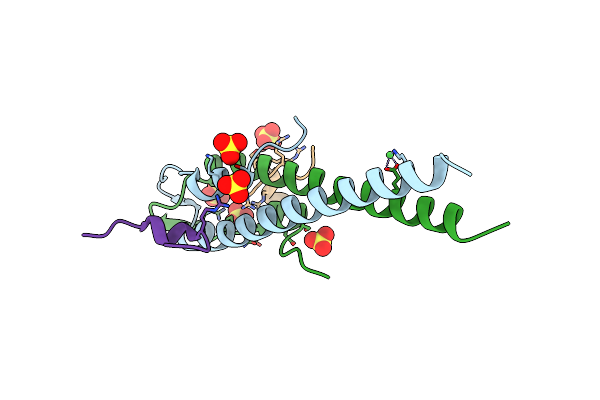 |
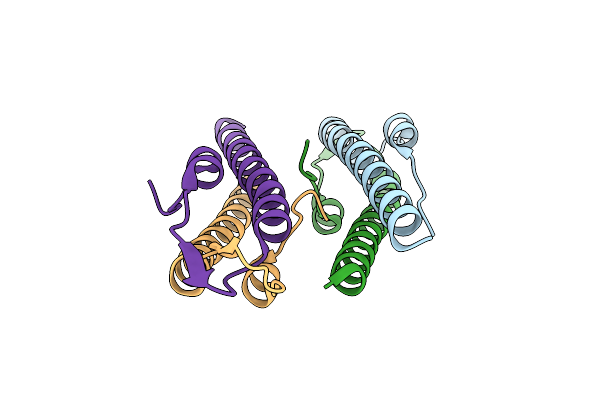
Cell Division Regulator, S. Pneumoniae Gpsb, In Complex With Peptide Fragment Of Penicillin Binding Protein Pbp2A
Organism: Streptococcus pneumoniae r6, Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-01-23 Classification: CELL CYCLE Ligands: SO4, NI |
 |
Crystal Structure Of The Streptococcus Pneumoniae D39 Copper Chaperone Cupa With Cu(I)
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2013-01-30 Classification: METAL TRANSPORT Ligands: CU1, CL |
 |
Crystal Structure Of The Metal Binding Domain (Mbd) Of The Streptococcus Pneumoniae D39 Cu(I) Exporting P-Type Atpase Copa With Cu(I)
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-01-30 Classification: METAL BINDING PROTEIN Ligands: CU1, CL |
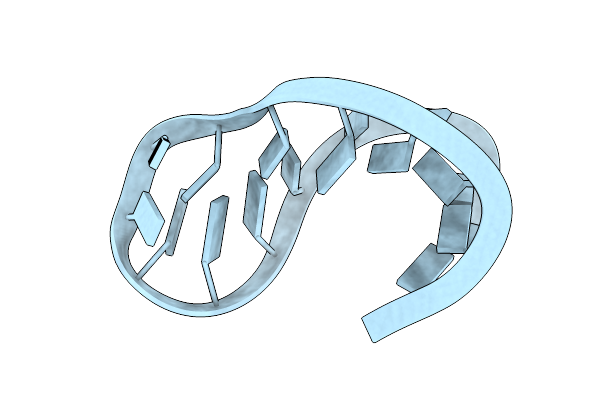 |
Solution Structure Of The Unmodified Anticodon Stem-Loop From E. Coli Trna(Phe)
|
 |
Solution Structure Of The Unmodified Anticodon Stem-Loop From E. Coli Trna(Phe)
|
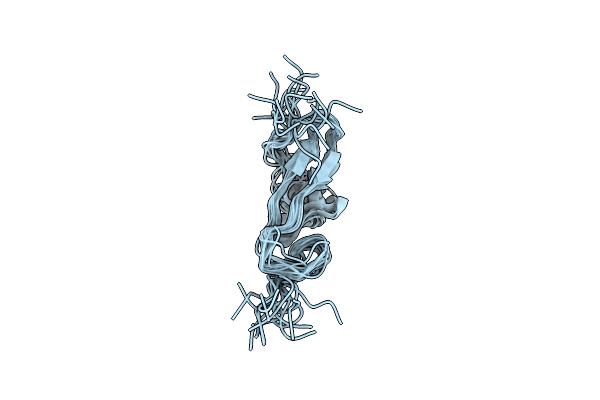 |
Type Alpha Transforming Growth Factor, Nmr, 16 Models Without Energy Minimization
|
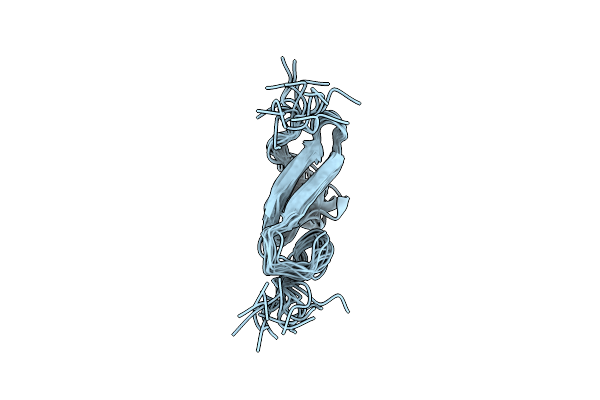 |
Type Alpha Transforming Growth Factor, Nmr, 15 Models After Ecepp/3 Energy Minimization
|

