Search Count: 15
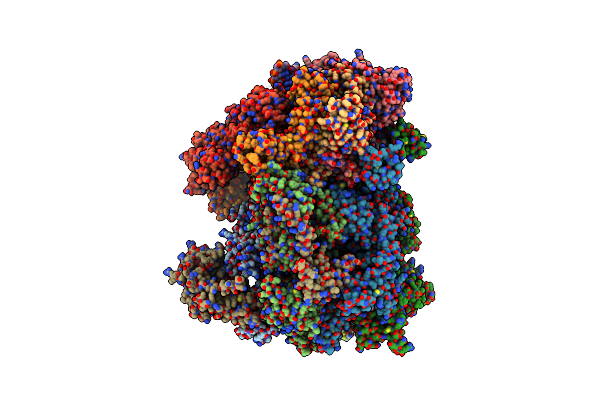 |
Cryo-Em Structure Of The S. Cerevisiae Orc-Cdc6-Mcm2-7-Dna Complex With A Fully Closed Mcm2-Mcm5 Dna Entry Gate
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: DNA BINDING PROTEIN Ligands: ATP, MG |
 |
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-03-08 Classification: OXIDOREDUCTASE Ligands: CL, SO4 |
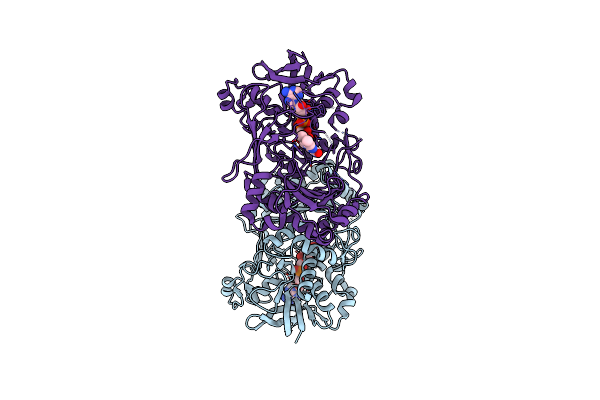 |
Crystal Structure Of The V465T Mutant Of 5-(Hydroxymethyl)Furfural Oxidase (Hmfo)
Organism: Methylovorus sp. (strain mp688)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-02-14 Classification: FLAVOPROTEIN Ligands: FAD |
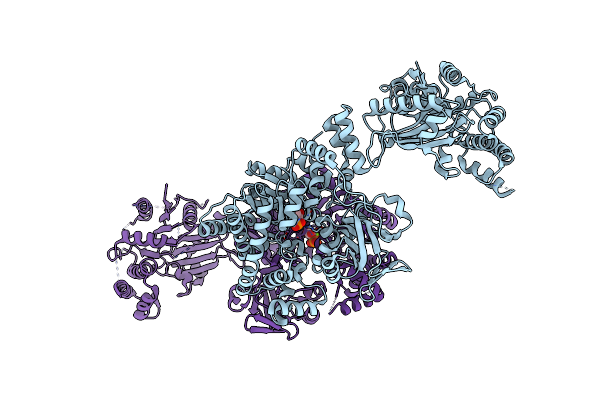 |
C4-Type Pyruvate Phosphate Dikinase: Different Conformational States Of The Nucleotide Binding Domain In The Dimer
Organism: Flaveria trinervia
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-04-05 Classification: TRANSFERASE Ligands: PEP, MG |
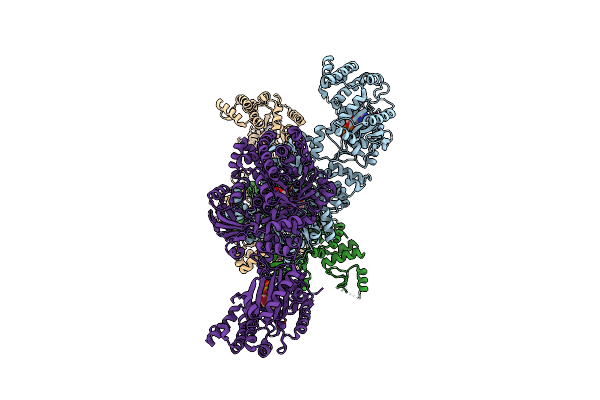 |
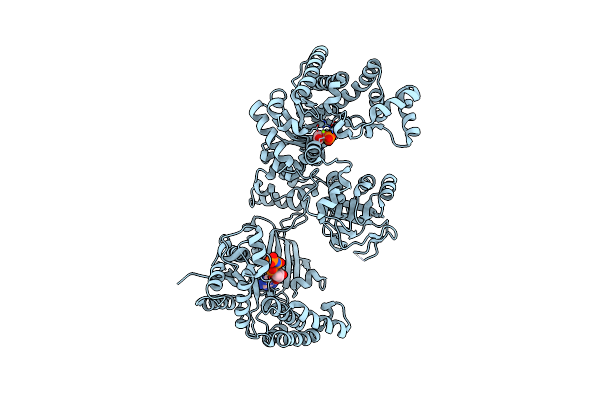
C4-Type Pyruvate Phospate Dikinase: Nucleotide Binding Domain With Bound Atp Analogue
Organism: Flaveria trinervia
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-04-05 Classification: TRANSFERASE Ligands: 6NQ, MG, PEP |
 |
C3-Type Pyruvate Phosphate Dikinase: Intermediate State Of The Central Domain In The Swiveling Mechanism
Organism: Flaveria pringlei
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-04-05 Classification: TRANSFERASE Ligands: 6NQ, PEP, MG |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound P-Hydroxybenzaldehyde
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HBA |
 |
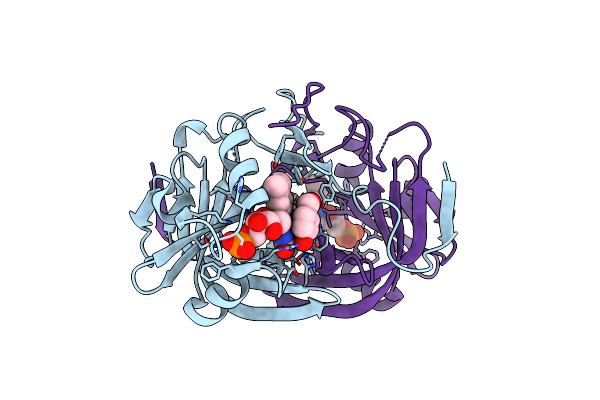
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound Benzene-1,4-Diol
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: FMN, HQE |
 |
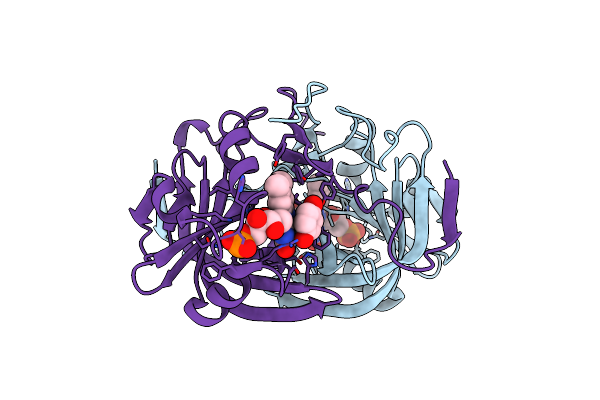
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound P-Hydroxybenzaldehyde
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HBA, FMN |
 |
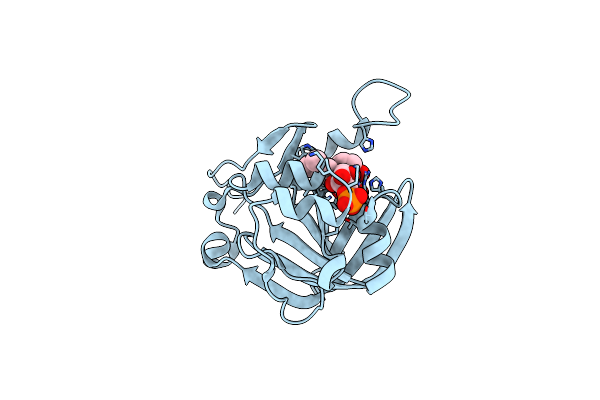
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound Benzene-1,4-Diol
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: HQE, FMN |
 |
Crystal Structure Of Fmn-Binding Protein (Np_142786.1) From Pyrococcus Horikoshii With Bound 1-Cyclohex-2-Enone
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-05-14 Classification: OXIDOREDUCTASE Ligands: FMN, A2Q |
 |
Crystal Structure Of Fmn-Binding Protein (Yp_005476) From Thermus Thermophilus With Bound 1-Cyclohex-2-Enone
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-05-14 Classification: FMN-BINDING PROTEIN Ligands: A2Q, FMN |
 |
 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-09-19 Classification: HYDROLASE Ligands: ZN, CA, PO4, NAG |

