Search Count: 26
 |
Structural Basis For The Autoinhibition And Sti-571 Inhibition Of C-Kit Tyrosine Kinase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-06-15 Classification: TRANSFERASE ACTIVATOR |
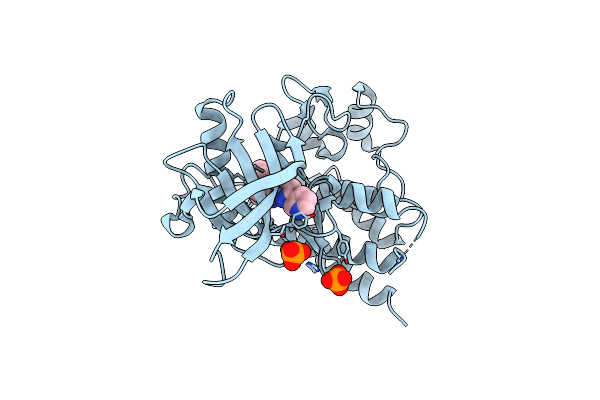 |
Structural Basis For The Autoinhibition And Sti-571 Inhibition Of C-Kit Tyrosine Kinase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-06-15 Classification: TRANSFERASE ACTIVATOR Ligands: PO4, STI |
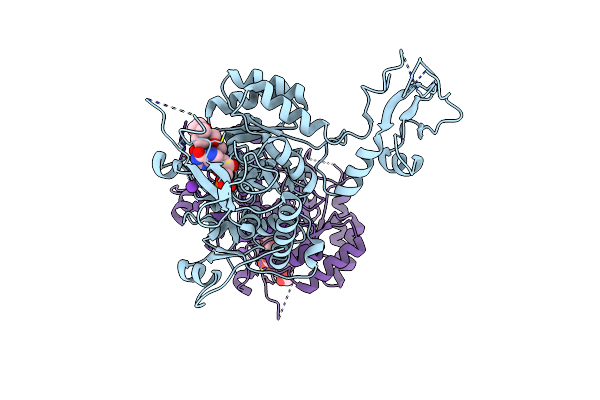 |
Crystal Structure Of Inosine Monophosphate Dehydrogenase In Complex With Mycophenolic Acid
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2001-09-05 Classification: OXIDOREDUCTASE Ligands: K, IMP, MOA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2001-05-23 Classification: APOPTOSIS, HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4 |
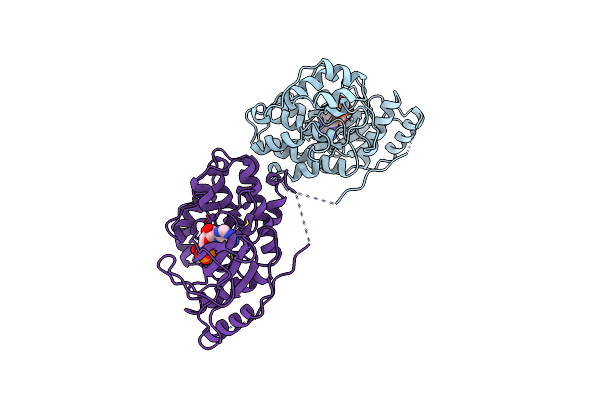 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2000-05-17 Classification: TRANSFERASE Ligands: MG, ANP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1999-06-08 Classification: TRANSFERASE Ligands: MG, ANP |
 |
Structure Of Penta Mutant Human Erk2 Map Kinase Complexed With A Specific Inhibitor Of Human P38 Map Kinase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-06-08 Classification: TRANSFERASE Ligands: SO4, SB2 |
 |
Structure Of Apo, Unphosphorylated, P38 Mitogen Activated Protein Kinase P38 (P38 Map Kinase) The Mammalian Homologue Of The Yeast Hog1 Protein
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1997-09-19 Classification: TRANSFERASE |
 |
Atomic Structure Of Fkbp12-Fk506, An Immunophilin Immunosuppressant Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1995-12-07 Classification: ROTAMASE Ligands: FK5 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1995-12-07 Classification: ROTAMASE Ligands: SO4 |
 |
Atomic Structure Of Fkbp12-Rapaymycin, An Immunophilin-Immunosuppressant Complex
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1995-12-07 Classification: ROTAMASE Ligands: RAP |
 |
Enhancement Of Protein Stability By The Combination Of Point Mutations In T4 Lysozyme Is Additive
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1995-07-31 Classification: HYDROLASE (O-GLYCOSYL) |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1995-07-28 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
The E. Coli Biotin Holoenzyme Synthetase(Slash)Bio Repressor Crystal Structure Delineates The Biotin And Dna-Binding Domains
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1993-10-31 Classification: TRANSCRIPTION REGULATION |
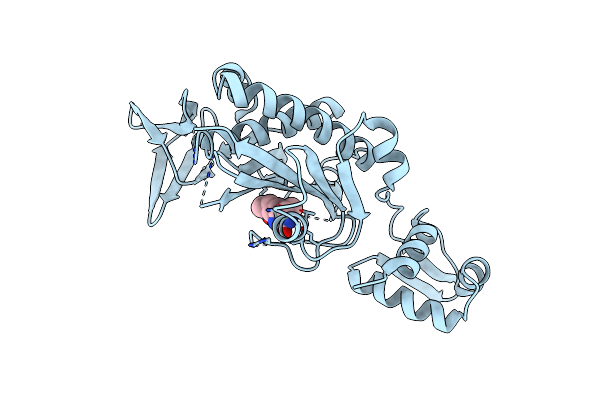 |
The E. Coli Biotin Holoenzyme Synthetase(Slash)Bio Repressor Crystal Structure Delineates The Biotin And Dna-Binding Domains
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1993-10-31 Classification: TRANSCRIPTION REGULATION Ligands: BTN |
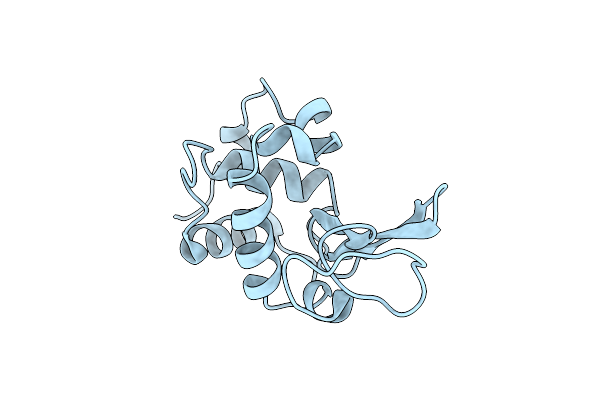 |
Structural And Thermodynamic Analysis Of Compensating Mutations Within The Core Of Chicken Egg White Lysozyme
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |
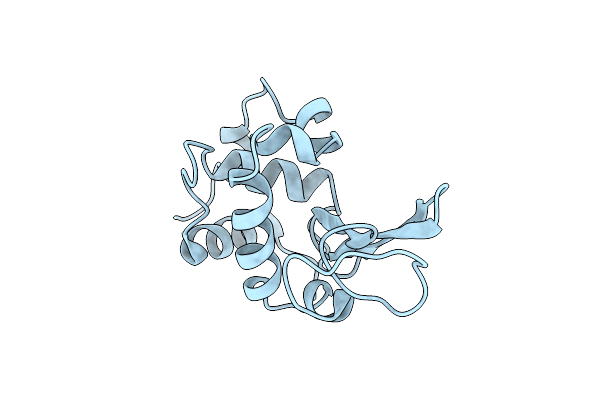 |
Structural And Thermodynamic Analysis Of Compensating Mutations Within The Core Of Chicken Egg White Lysozyme
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |
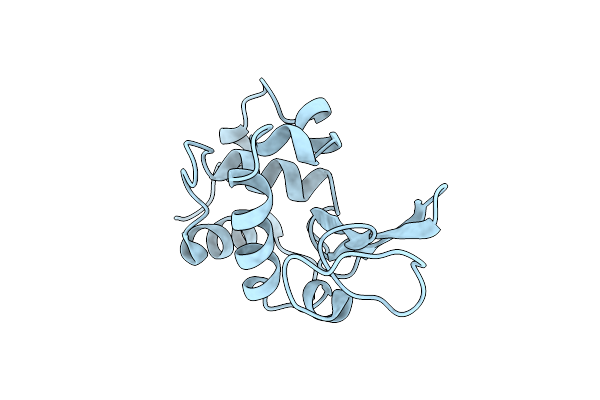 |
Structural And Thermodynamic Analysis Of Compensating Mutations Within The Core Of Chicken Egg White Lysozyme
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |
 |
Structural And Thermodynamic Analysis Of Compensating Mutations Within The Core Of Chicken Egg White Lysozyme
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |
 |
Structural And Thermodynamic Analysis Of Compensating Mutations Within The Core Of Chicken Egg White Lysozyme
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |

