Search Count: 21
 |
Co-Structure Of The Fab Of The Anti-Tigit Vibostolimab Antibody With Its Antigen
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Cryoem Structure Of The F Plasmid Relaxosome In Its Pre-Initiation State, Derived From The Ds-27_+143-R Locally-Refined Map 3.76 A
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Trai In Its Te Mode, Derived From The Ss-27_+8Ds+9_+143-R Locally-Refined 3.45 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Truncated Trai1-863 In Its Te Mode, Derived From The Ss-27_+8Ds+9_+143-R_Deltaah+Ctd Locally-Refined 3.42 A Map
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Trai In Its Te Mode, Without Accessory Protein Tram. Derived From The Ss-27_+8Ds+9_+143-R_Deltatram Locally-Refined 2.94 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Orit Dna Ss-27_+3Ds+4_+143 And Trai Its Te Mode, Derived From Ss-27_+3Ds+4_+143-R Locally-Refined 3.68 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Cryoem Structure Of The F Plasmid Relaxosome With Orit Dna Ss-27_-3Ds-2_+143 And Trai Its Te Mode, Derived From Ss-27_-3Ds-2_+143-R Locally-Refined 3.42 A Map.
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Co-Structure Of The Fab Of The Anti-Tigit Vibostolimab Antibody With Its Antigen
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-05-28 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2021-04-21 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: Y5Y, EDO, SO4 |
 |
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: FE2 |
 |
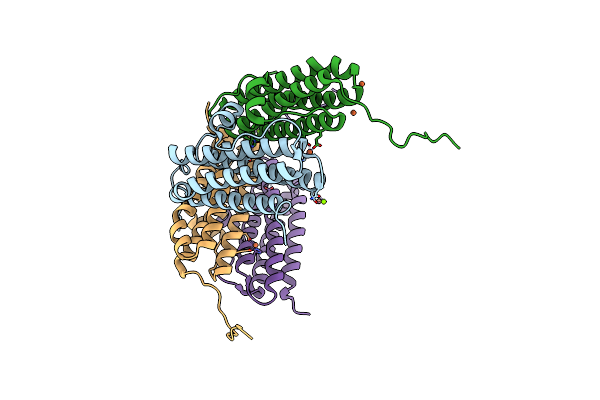
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 24 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, FE2, MG, CL |
 |
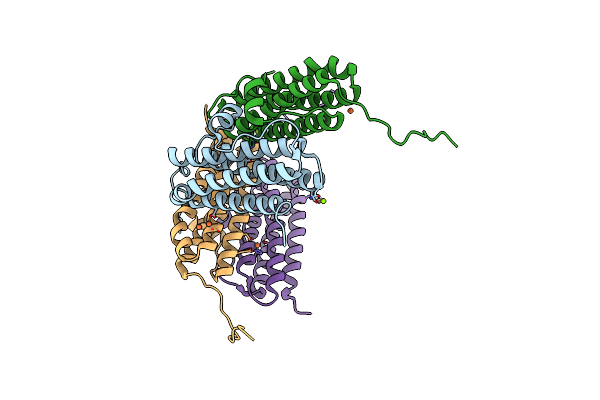
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 48 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, MG, CL, FE2 |
 |
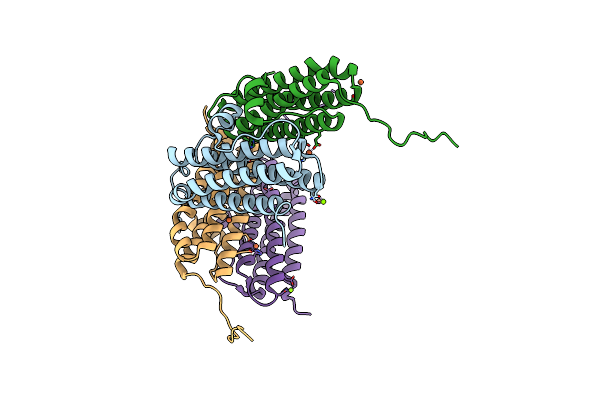
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 100 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, FE2, MG, CL |
 |
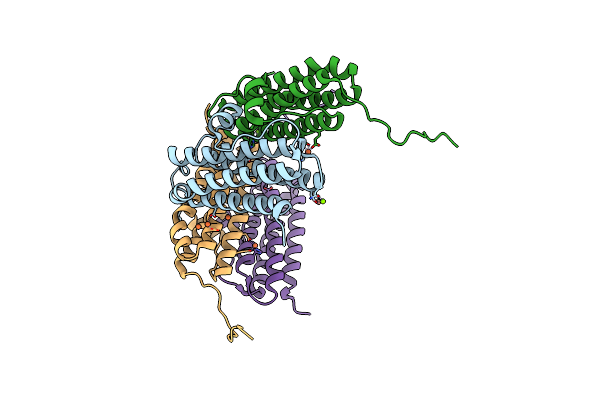
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 240 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, FE2, MG, CL |
 |
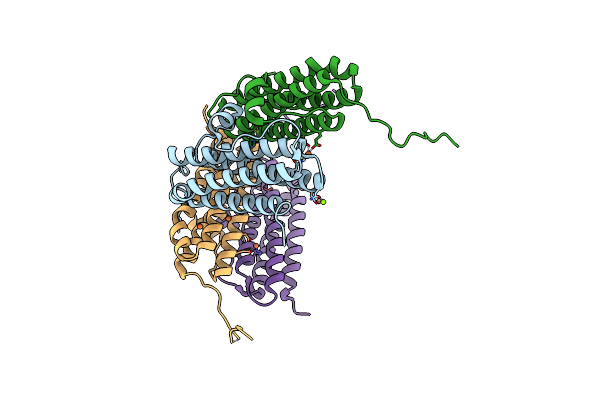
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 360 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, FE2, MG, CL |
 |
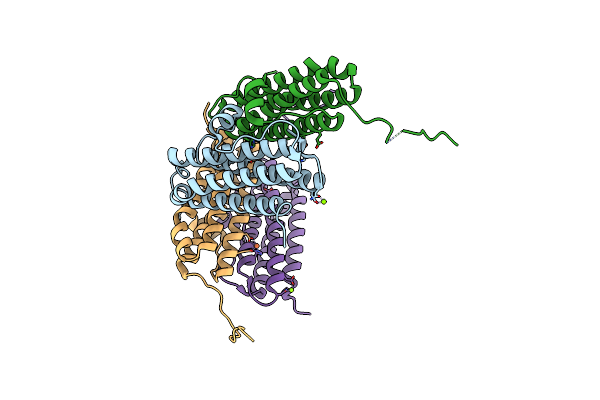
Crystal Structure Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis Soaked With Iron In The Ratio Of 480 Iron Atoms Per Dodecamer
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: FE, FE2, MG, CL |
 |
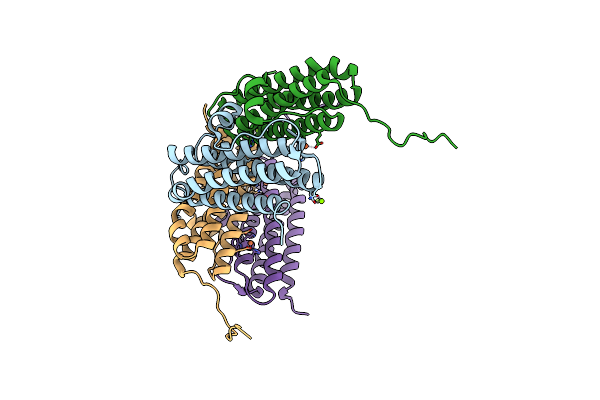
Crystal Structure Of R73E Mutant Of The Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-09 Classification: DNA BINDING PROTEIN Ligands: MG, CL, FE |
 |
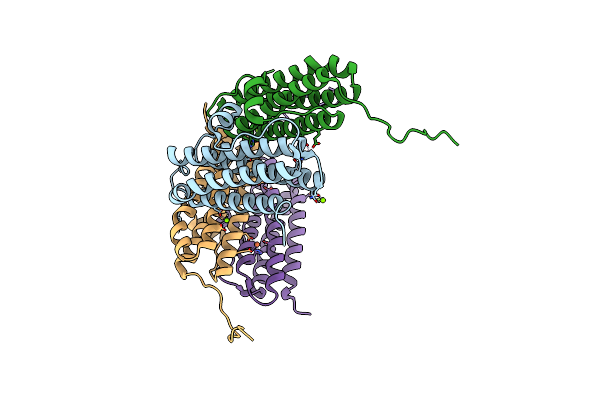
Crystal Structure Of Gated-Pore Mutant D138N Of Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2014-03-05 Classification: DNA BINDING PROTEIN Ligands: MG, CL, FE2 |
 |
Crystal Structure Of Gated-Pore Mutant H141D Of Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2014-03-05 Classification: DNA BINDING PROTEIN Ligands: FE2, MG, CL |
 |
Crystal Structure Of Gated-Pore Mutant D138H Of Second Dna-Binding Protein Under Starvation From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-03-05 Classification: DNA BINDING PROTEIN Ligands: FE2, MG, CL |

