Search Count: 232
 |
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Crystal Structure Of Gh139 Glycoside Hydrolase From Verrucomicrobium Sp. In The Hexagonal Space Group P6522
Organism: Verrucomicrobium sp.
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE, NA, CL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium (Unmodelled Additional Ligand Density At Active Site)
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE |
 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Nad+ (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Sulfoquinovose Substrate (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: YZT |
 |
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: EDO, ZN |
 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn From Cupriavidus Pinatubonensis In Complex With Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI |
 |
Crystal Structure Of Hpsn D352A Mutant From Cupriavidus Pinatubonensis In Complex With Nad+
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn From Cupriavidus Pinatubonensis In Complex With Product Analogue (L-Cysteate) And Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, OCS |
 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn From Cupriavidus Pinatubonensis In Complex With Product (R-Sulfolactate) And Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, 3SL |
 |
Crystal Structure Of Dhps-3-Dehydrogenase, Hpsn H319A Variant From Cupriavidus Pinatubonensis In Complex With Substrate (R-Dhps) And Nadh
Organism: Cupriavidus pinatubonensis jmp134
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-09-25 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, A1AZH, SO4 |
 |
Crystal Structure Of Sulfohexulose-1-Phosphate Aldolase From Paracoccus Onubensis Strain Merri
Organism: Paracoccus onubensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-08-07 Classification: LYASE |
 |
Structure Of An Allelic Variant Of Puccinia Graminis F. Sp. Tritici (Pgt) Effector Avrsr27 (Avrsr27-1)
Organism: Puccinia graminis f. sp. tritici
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-06-12 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
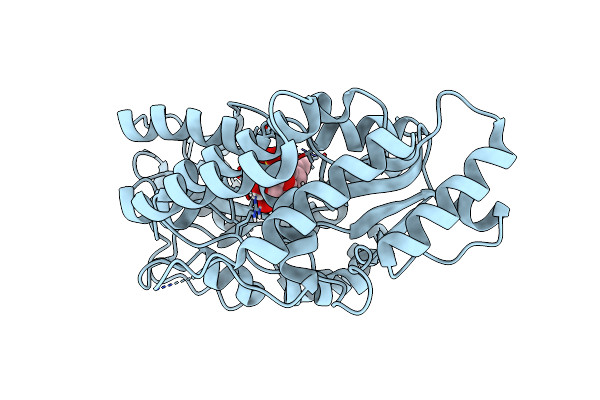
Crystal Structure Of Gh31 Family Sulfoquinovosidase Bmsqase In Covalent Complex With Sq-Aziridine (Sqz)
Organism: Priestia megaterium dsm 319
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-05-08 Classification: HYDROLASE Ligands: Y2W, K |
 |
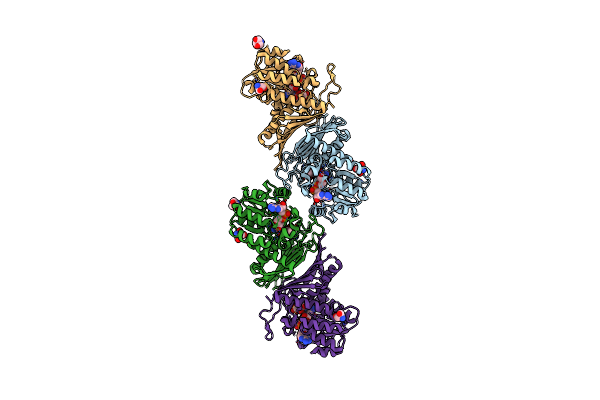
Crystal Structure Of The Sulfoquinovosyl Binding Protein (Smof) From A. Tumefaciens Sulfo-Smo Pathway In Complex With Sqoctyl Ligand
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-04-17 Classification: SUGAR BINDING PROTEIN Ligands: A1H5E |
 |
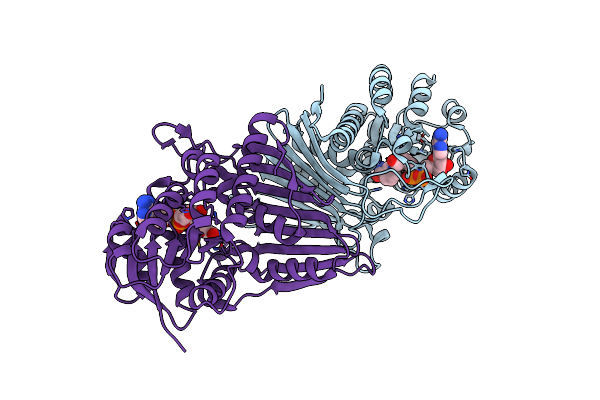
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Flavobacterium Sp. (Strain K172) In Complex With Co-Factor Nad+ And Sulfoquinovose (Sq)
Organism: Paenarthrobacter ureafaciens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD, R7R, TRS |
 |
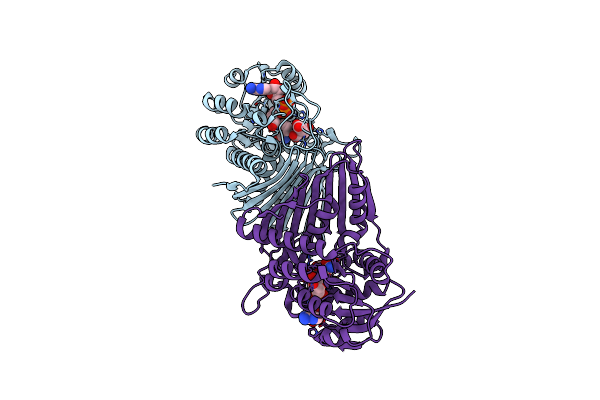
Crystal Structure Of Oxidoreductive Sulfoquinovosidase From Arthrobacter Sp. U41 (Arsqga)In Complex With Co-Factor Nad+
Organism: Arthrobacter sp. u41
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD |
 |
Crystal Structure Of Nad-Dependent Glycoside Hydrolase From Arthrobacter Sp. U41 In Complex With Nad+ Cofactor And Citrate
Organism: Arthrobacter sp. u41
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-12-27 Classification: HYDROLASE Ligands: NAD, CIT |

