Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
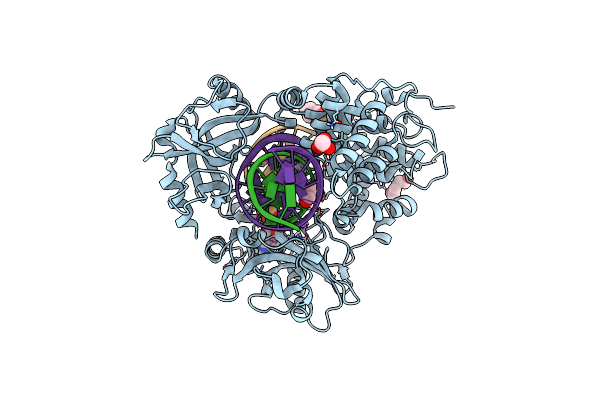 |
Human Dna Ligase I F872A Bound To Adenylated Nicked Dna With A 5' Terminal Ribonucleotide
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LIGASE/DNA Ligands: PEG, MES, CL, GOL, NA, AMP |
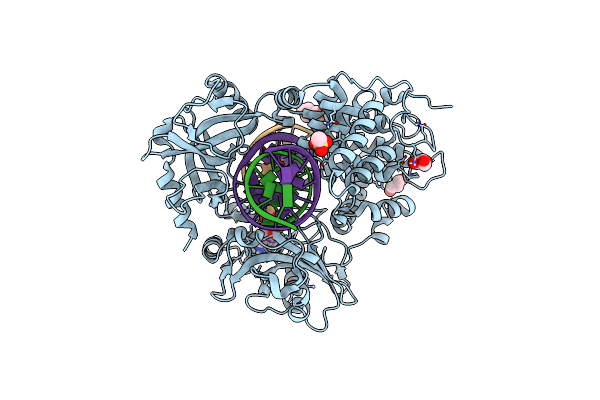 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LIGASE/DNA Ligands: GOL, MES, AMP, NA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LIGASE/DNA Ligands: MES, AMP |
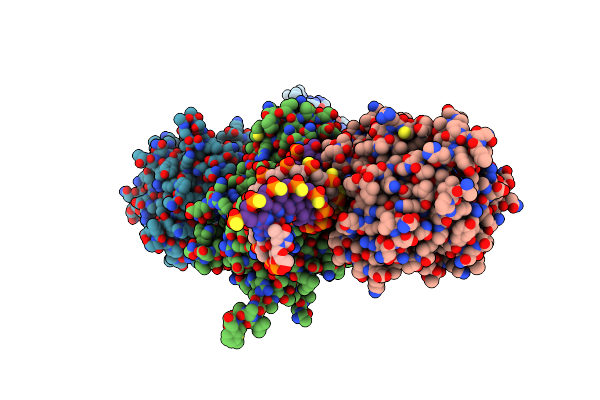 |
Crystal Structure Of Saccharomyces Cerevisiae Apn2 Catalytic Domain E59Q/D222N Mutant In Complex With Dna
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2022-09-28 Classification: HYDROLASE/DNA Ligands: TAR, CA, TLA, CIT, EDO |
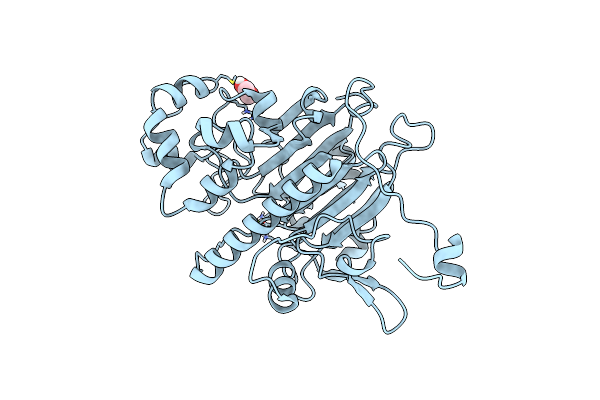 |
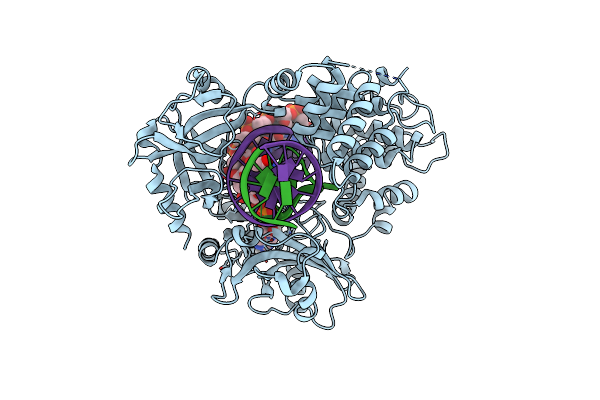
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: MG, CL, EDO |
 |
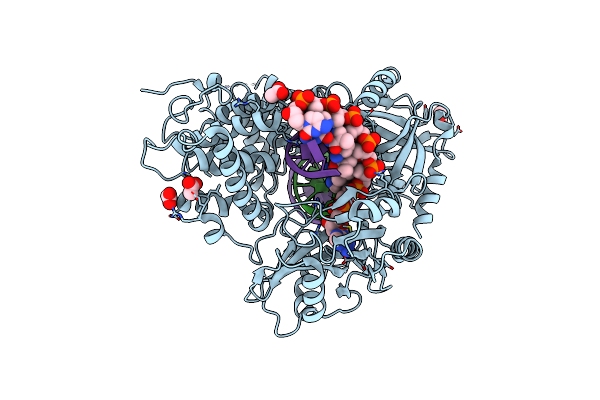
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2021-02-03 Classification: LIGASE/DNA Ligands: AMP, ACT |
 |
Human Dna Ligase 1(E346A/E592A) Bound To A Nicked Dna Substrate Control Duplex
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-02-03 Classification: LIGASE/DNA Ligands: ACT, AMP |
 |
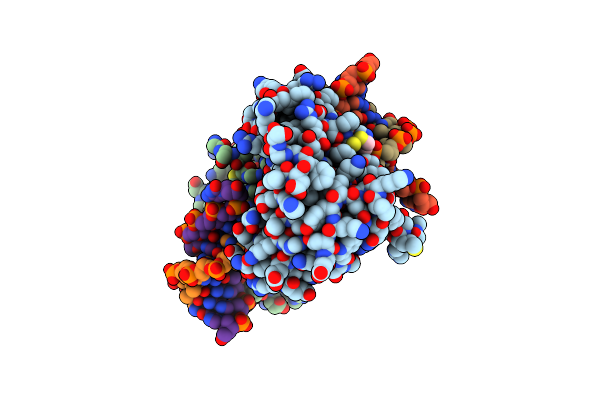
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-01-14 Classification: DNA BINDING PROTEIN Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2013-12-18 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: AMP, ZN, NA, BME, K |
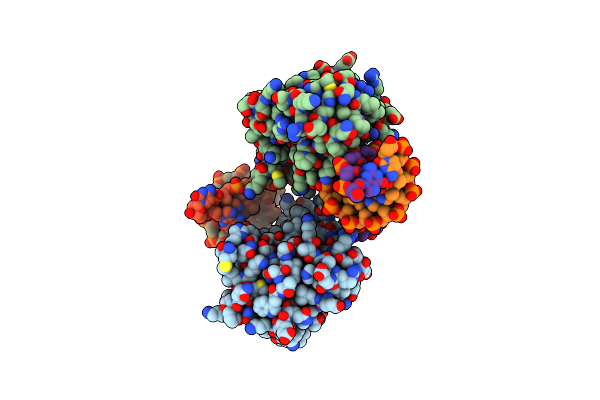 |
Human Aprataxin (Aptx) Bound To Rna-Dna And Zn - Adenosine Vanadate Transition State Mimic Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2013-12-18 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: V5A, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-12-18 Classification: DNA BINDING PROTEIN/DNA Ligands: AMP, ZN |
 |
Human Aprataxin (Aptx) Aoa1 Variant K197Q Bound To Rna-Dna, Amp, And Zn - Product Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-12-18 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: AMP, ZN, NA, BME, GOL, K |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2011-10-12 Classification: HYDROLASE/DNA Ligands: AMP, BME, ZN |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2011-03-30 Classification: REPLICATION Ligands: PO4 |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-03-30 Classification: REPLICATION |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-03-30 Classification: REPLICATION Ligands: ANP, MG |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2011-03-30 Classification: REPLICATION Ligands: ANP, MG |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2010-02-09 Classification: CELL CYCLE |
 |
Crystal Structure Of S. Pombe Brc1 Brct5-Brct6 Domains In Complex With Phosphorylated H2A
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2010-02-09 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2009-10-13 Classification: CELL CYCLE |