Search Count: 91
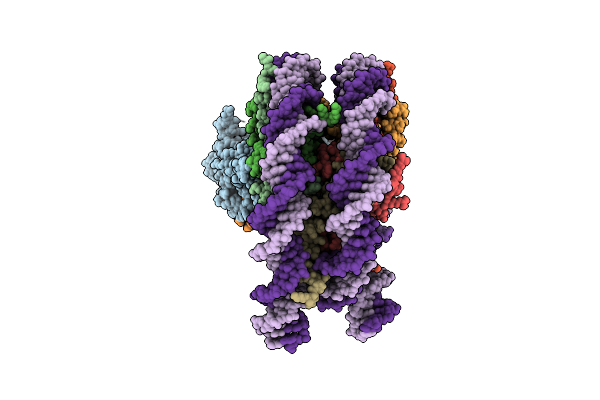 |
Organism: Arabidopsis thaliana, Homo sapiens, Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: TRANSLOCASE Ligands: ADP, BEF |
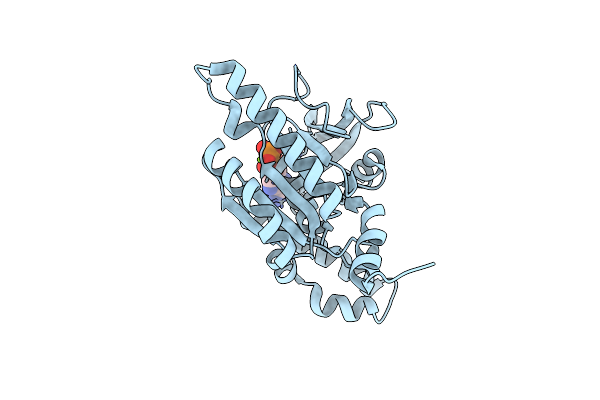 |
Organism: Alvinella pompejana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-08-16 Classification: DNA BINDING PROTEIN Ligands: MG, ADP |
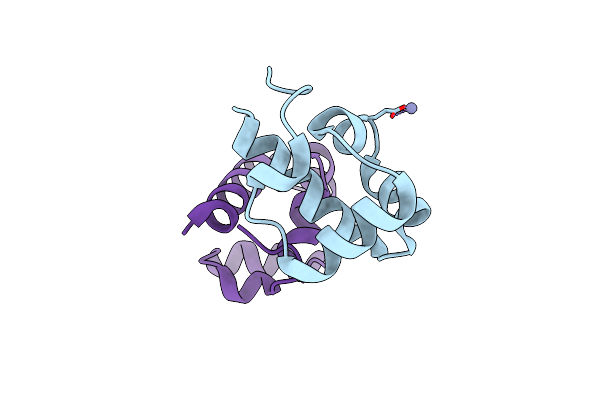 |
Organism: Alvinella pompejana
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-16 Classification: DNA BINDING PROTEIN Ligands: ZN |
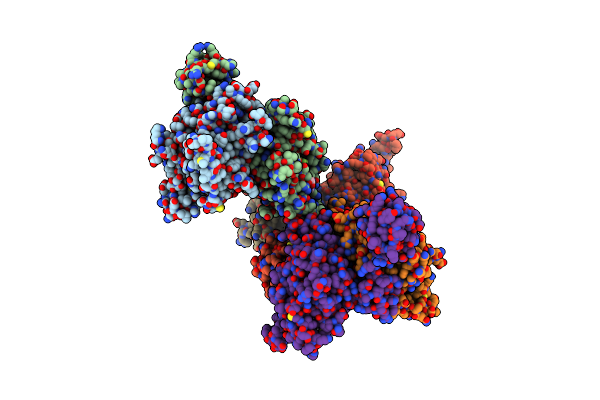 |
Organism: Alvinella pompejana
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-08-16 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 1 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 20 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 8 Us Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Directed Evolution Of A Biosensor Selective For The Macrolide Antibiotic Clarithromycin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-19 Classification: DNA BINDING PROTEIN/ANTIBIOTIC Ligands: CTY, FLC |
 |
In Cellulo Crystallization Of Trypanosoma Brucei Imp Dehydrogenase Enables The Identification Of Atp And Gmp As Genuine Co-Factors
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-02-19 Classification: IMMUNOSUPPRESSANT Ligands: ATP, 5GP |
 |
Structure Of Native Granulovirus Polyhedrin Determined Using An X-Ray Free-Electron Laser
Organism: Cydia pomonella granulovirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-02-22 Classification: STRUCTURAL PROTEIN |
 |
Organism: Cydia pomonella granulovirus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2017-02-22 Classification: VIRAL PROTEIN |
 |
Organism: Lysinibacillus sphaericus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-10-05 Classification: TOXIN |
 |
Organism: Lysinibacillus sphaericus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-10-05 Classification: TOXIN |
 |
Organism: Lysinibacillus sphaericus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-10-05 Classification: TOXIN |
 |
Bacteriorhodopsin Ground State Structure Obtained With Serial Femtosecond Crystallography
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-08-31 Classification: PROTON TRANSPORT Ligands: RET, LI1 |
 |
Organism: Enterobacteria phage t4, Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:7.70 Å Release Date: 2016-03-23 Classification: SIGNALING PROTEIN |

