Search Count: 20
 |
Ttx183A - A C-Type Cytochrome Domain From The Teredinibacter Turnerae Protein Tertu_2913
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-03-13 Classification: ELECTRON TRANSPORT Ligands: HEC, CL |
 |
Ttx183B - A C-Type Cytochrome Domain From The Teredinibacter Turnerae Protein Tertu_2913
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: ELECTRON TRANSPORT Ligands: HEC, CL, NA |
 |
Se-Met Labelled Ttx122A - A Domain Of Unknown Function From The Teredinibacter Turnerae Protein Tertu_3803
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-03-13 Classification: UNKNOWN FUNCTION Ligands: CA, MG, EDO |
 |
Ttx122A - A Domain Of Unknown Function From The Teredinibacter Turnerae Protein Tertu_3803
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-03-13 Classification: UNKNOWN FUNCTION Ligands: CA, MG, EDO |
 |
Ttx122B - A Domain Of Unknown Function From The Teredinibacter Turnerae Protein Tertu_2913
Organism: Teredinibacter turnerae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-13 Classification: UNKNOWN FUNCTION Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2018-12-19 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Lactobacillus reuteri atcc 53608
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2018-03-21 Classification: CELL ADHESION |
 |
Organism: Lactobacillus reuteri 100-23
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-03-21 Classification: CELL ADHESION |
 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN Ligands: CA, NA |
 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1 In The Orthorhombic Form
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: 2PE, P6G, EDO, GOL, PEG, CA, PG4, HHD |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1 Collected At The Zn Edge
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1 At Medium Resolution
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
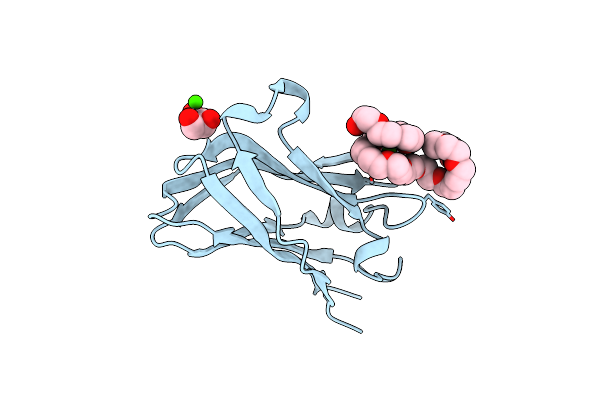 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: 2PE, P6G, CA, BGQ |
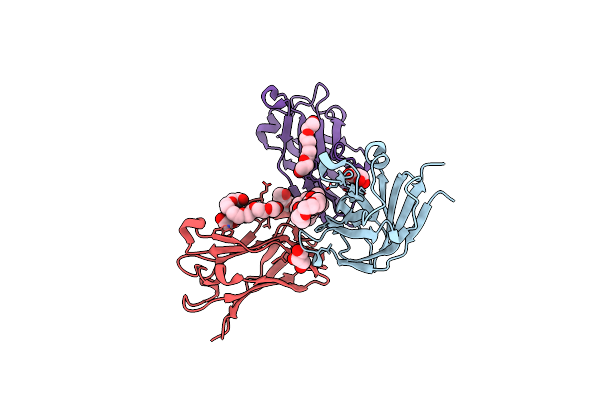 |
High Resolution Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: P6G, GOL, PGE, PG4 |
 |
Atomic Resolution Structure Of Tmcbm61 In Complex With Beta-1,4- Galactotriose
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2010-09-08 Classification: HYDROLASE Ligands: CA |
 |
Structure Of Tmcbm61 In Complex With Beta-1,4-Galactotriose At 1.4 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-09-08 Classification: HYDROLASE Ligands: CA, SO4, EDO |

