Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
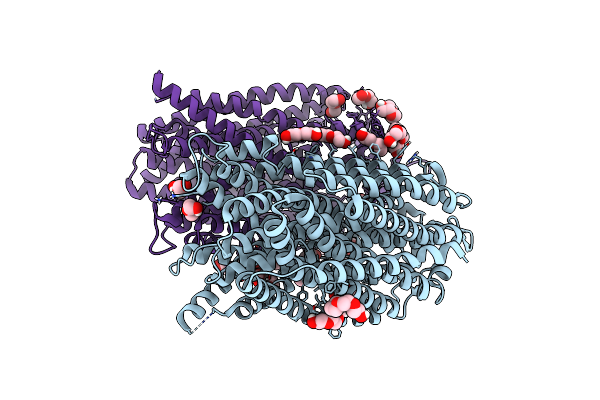 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 0-60-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: LMT, PEG, PGE, PG4 |
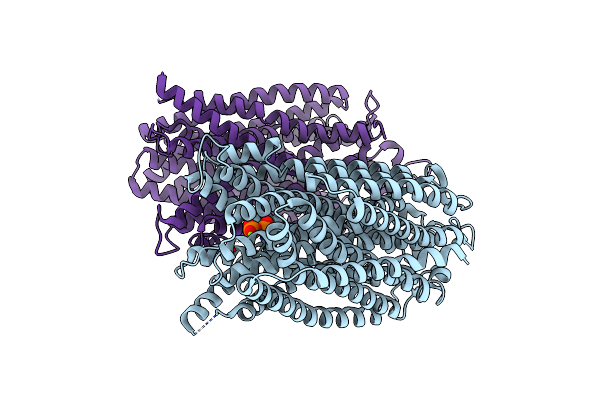 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 300-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.98 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG |
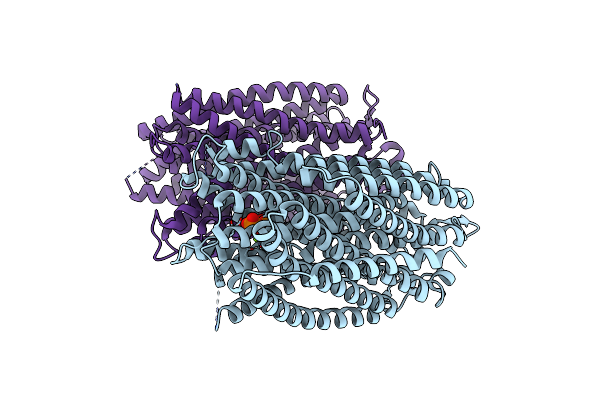 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 600-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG |
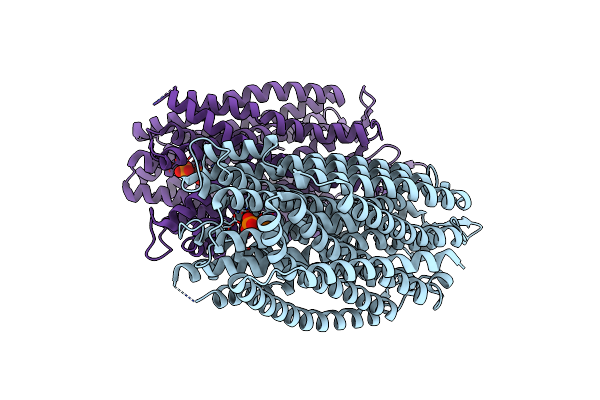 |
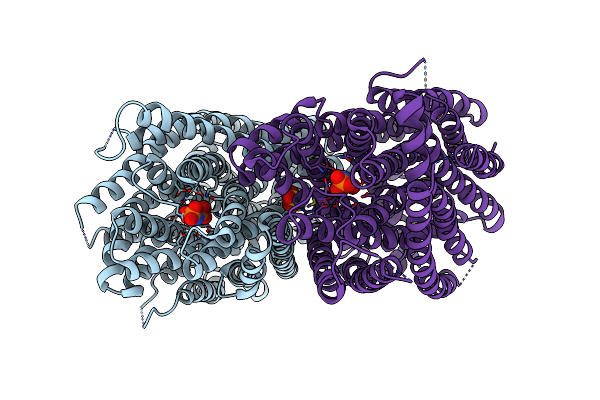
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 3600-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:4.53 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG, K, PO4 |
 |
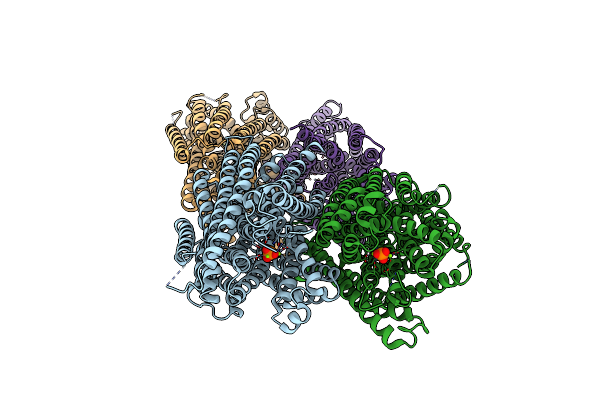
Crystal Structure Of Pyrobaculum Aerophilum Potassium-Independent Proton Pumping Membrane Integral Pyrophosphatase In Complex With Imidodiphosphate And Magnesium, And With Bound Sulphate
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: MG, 2PN, SO4 |
 |
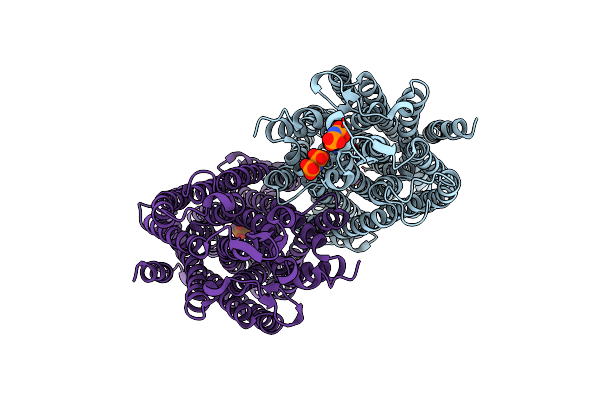
Organism: Vigna radiata var. radiata
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-12-28 Classification: HYDROLASE Ligands: PO4, MG |
 |
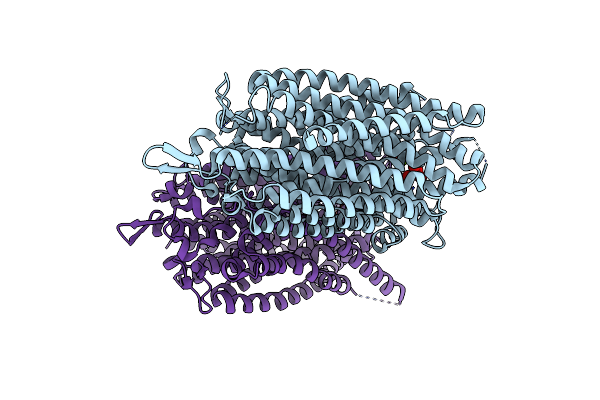
Crystal Structure Of Thermotoga Maritima Sodium Pumping Membrane Integral Pyrophosphatase In Complex With Imidodiphosphate And Magnesium, And With Bound Sodium Ion
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: MG, 2PN, NA |
 |
Crystal Structure Of Thermotoga Maritima Sodium Pumping Membrane Integral Pyrophosphatase In Complex With Tungstate And Magnesium
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: WO4, MG |
 |
Organism: Toxoplasma gondii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-02-05 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of Hepatitis C Virus Polymerase In Complex With Inhibitor Sb655264
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-02-13 Classification: HYDROLASE Ligands: 699 |
 |
Crystal Structure Of Hepatitis C Virus Polymerase In Complex With Inhibitor Sb698223
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-02-13 Classification: HYDROLASE Ligands: 698 |
 |
Hydrogen And Hydration Of Hen Egg-White Lysozyme Determined By Neutron Diffraction
Organism: Gallus gallus
Method: NEUTRON DIFFRACTION Resolution:2.00 Å Release Date: 2001-02-07 Classification: HYDROLASE Ligands: DOD |
 |
Direct Determination Of The Positions Of Deuterium Atoms Of Bound Water In Concanavalin A By Neutron Laue Crystallography
Organism: Canavalia ensiformis
Method: NEUTRON DIFFRACTION Resolution:2.40 Å Release Date: 2000-05-08 Classification: SUGAR BINDING PROTEIN Ligands: MN, CA |
 |
Organism: Canavalia ensiformis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2000-04-30 Classification: LECTIN Ligands: MN, CA |
 |
Organism: Gallus gallus
Method: NEUTRON DIFFRACTION Resolution:1.70 Å Release Date: 1999-04-01 Classification: HYDROLASE Ligands: NO3, NA, DOD |