Search Count: 27
 |
Human V-Atpase Vo Subcomplex (Containing Subunit Isoform A4) Bound To Nanobody And Inhibitor
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN Ligands: NAG, CLR, POV, A1A4Q, WJP |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
 |
Organism: Lama glama, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2025-02-05 Classification: PROTON TRANSPORT |
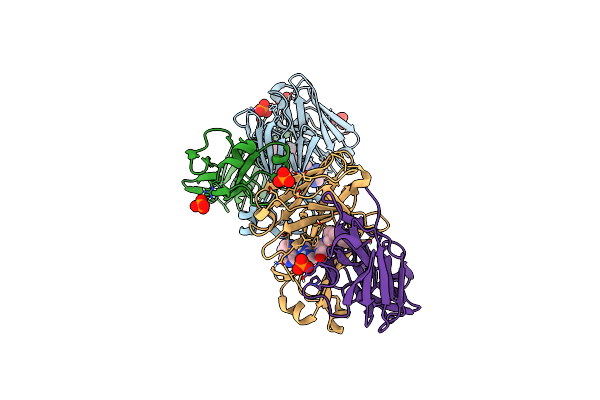 |
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 1-(1-(Imidazo[1,5-A]Pyrazin-8-Yl)Azetidin-3-Yl)-3-(2-(6-Methyl-4-(3-Methyl-3-Phenylpyrrolidin-1-Yl)-2-Oxopyridin-1(2H)-Yl)Ethyl)Urea
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: PO4, A1AHP |
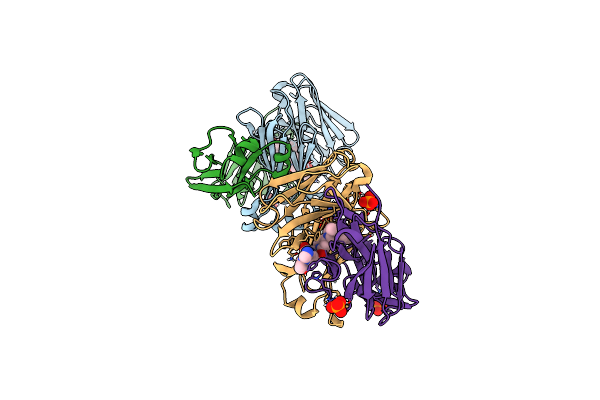 |
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 3-(2-(4-(3-Ethyl-3-Phenylpyrrolidin-1-Yl)-6-Methyl-2-Oxopyridin-1(2H)-Yl)Ethyl)-8-(Imidazo[1,5-A]Pyrazin-8-Yl)-1,3,8-Triazaspiro[4.5]Decan-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: PO4, A1AH0 |
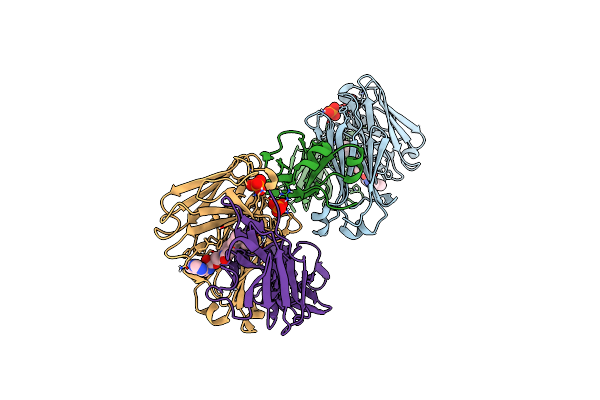 |
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 2-(6-Methyl-2-Oxo-4-(2-Phenylpropoxy)Pyridin-1(2H)-Yl)Ethyl (3-(1H-Imidazol-4-Yl)Benzyl)Carbamate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: PO4, A1AH2 |
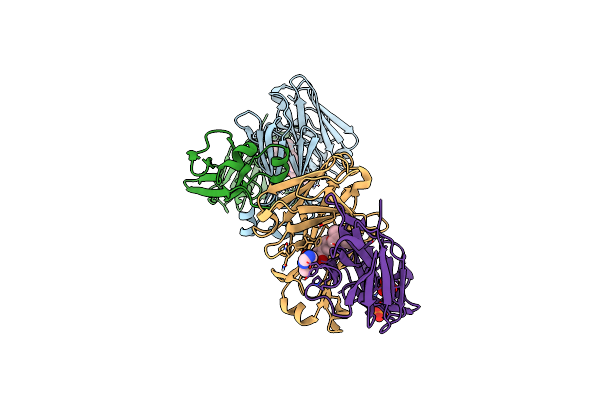 |
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 2-(1-Methyl-3-Oxo-5-(2-Phenylpropoxy)Isoindolin-2-Yl)Ethyl (3-(1H-Imidazol-4-Yl)Benzyl)Carbamate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: A1AH6, PO4 |
 |
Structure Of Full-Length Amyloidogenic Immunoglobulin Light Chain H9 In Complex With 4-(3-Ethyl-3-Phenylpyrrolidin-1-Yl)-1,6-Dimethylpyridin-2(1H)-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: PO4, A1AH1 |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2021-12-29 Classification: MOTOR PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MOTOR PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MOTOR PROTEIN |
 |
Organism: Saccharomyces cerevisiae s288c, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MOTOR PROTEIN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-11-04 Classification: TRANSPORT PROTEIN Ligands: PEE, PPV, EYR |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2020-11-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2018-03-21 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-06-21 Classification: Oxidoreductase/Inhibitor Ligands: SO4, 8UY, FE2 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2017-06-21 Classification: Oxidoreductase/Inhibitor Ligands: SO4, FE, 8UY |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:7.00 Å Release Date: 2016-06-08 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:6.20 Å Release Date: 2016-06-08 Classification: HYDROLASE |

