Search Count: 39
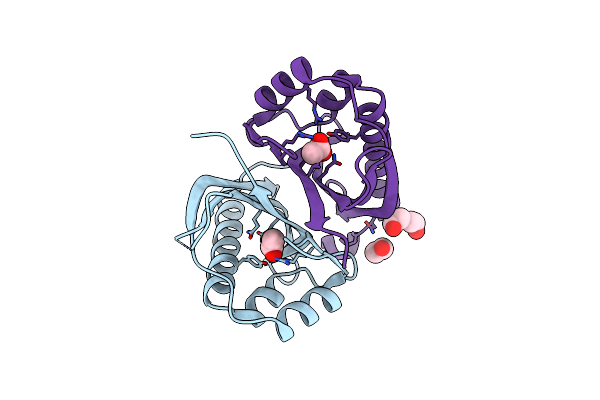 |
The Structure Of Atzh: A Little Known Member Of The Atrazine Breakdown Pathway
Organism: Pseudomonas sp. egd-akn5
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-11-14 Classification: UNKNOWN FUNCTION Ligands: ACY, PEG, EDO |
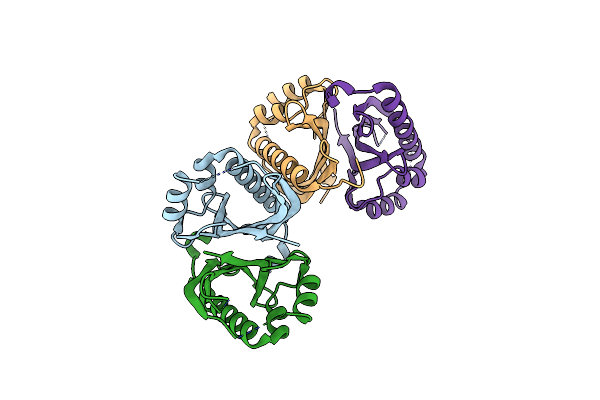 |
The Structure Of Atzh: A Little Known Member Of The Atrazine Breakdown Pathway
Organism: Pseudomonas sp. egd-akn5
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2018-11-14 Classification: UNKNOWN FUNCTION |
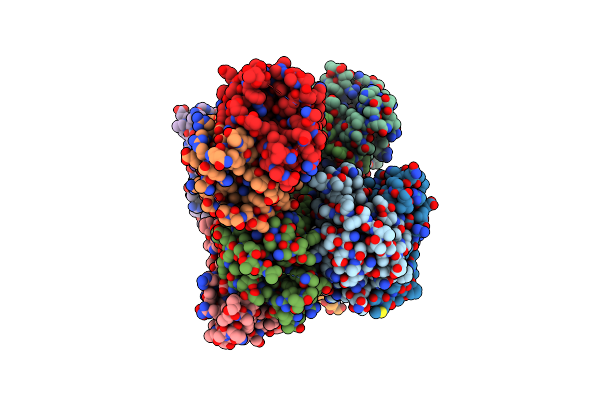 |
The Structure Of Atzh: A Little Known Member Of The Atrazine Breakdown Pathway
Organism: Pseudomonas sp. (strain adp)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: UNKNOWN FUNCTION Ligands: 6JN |
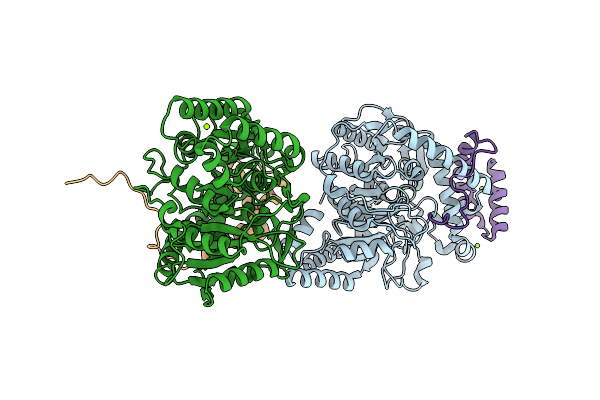 |
An Unexpected Vestigial Protein Complex Reveals The Evolutionary Origins Of An S-Triazine Catabolic Enzyme.
Organism: Pseudomonas sp. (strain adp)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: MG |
 |
An Unexpected Vestigial Protein Complex Reveals The Evolutionary Origins Of An S-Triazine Catabolic Enzyme. Inhibitor Bound Complex.
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: CA |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: BTB, GOL |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: EDO, PO4 |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: CL |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: C5J, CA |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-02-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: BTB, GOL, CL, C5S, PEG |
 |
Directed Evolution Of Transaminases By Ancestral Reconstruction. Using Old Proteins For New Chemistries
Organism: Pseudomonas sp. aac
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-07-12 Classification: TRANSFERASE Ligands: SO4, PLP, GOL, CL |
 |
Directed Evolution Of Transaminases By Ancestral Reconstruction. Using Old Proteins For New Chemistries
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2017-07-12 Classification: TRANSFERASE Ligands: PLP |
 |
Directed Evolution Of Transaminases By Ancestral Reconstruction. Using Old Proteins For New Chemistries
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2017-07-12 Classification: TRANSFERASE Ligands: MG, PEG, EDO |
 |
Directed Evolution Of Transaminases By Ancestral Reconstruction. Using Old Proteins For New Chemistries
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-07-12 Classification: TRANSFERASE Ligands: PLP |
 |
Directed Evolution Of Transaminases By Ancestral Reconstruction. Using Old Proteins For New Chemistries
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-07-12 Classification: TRANSFERASE Ligands: PLP, EDO |
 |
Directed Evolution Of Transaminases By Ancestral Reconstruction. Using Old Proteins For New Chemistries
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-07-12 Classification: TRANSFERASE Ligands: PLP, CA, PEG |
 |
Directed Evolution Of Transaminases By Ancestral Reconstruction. Using Old Proteins For New Chemistries
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-07-12 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: MG, SO4, MHA |
 |
Structure-Function Analysis Of Functionally Diverse Members Of The Cyclic Amide Hydrolase Family Of Toblerone Fold Enzymes
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: SO4, MHA, NA, CL |
 |
Structure-Function Analysis Of Functionally Diverse Members Of The Cyclic Amide Hydrolase Family Of Toblerone Fold Enzymes
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2017-02-01 Classification: HYDROLASE Ligands: CL, MHA, NA |

