Search Count: 53
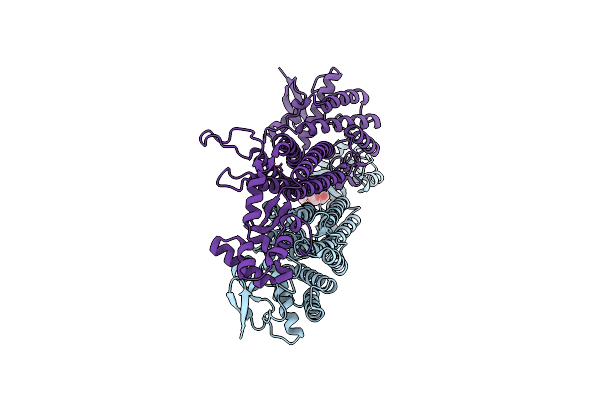 |
Crystal Structure Of Mutant Aspartase From Bacillus Sp. Ym55-1 In The Closed Loop Conformation
Organism: Bacillus sp. ym55-1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: LYASE Ligands: PGE, NA |
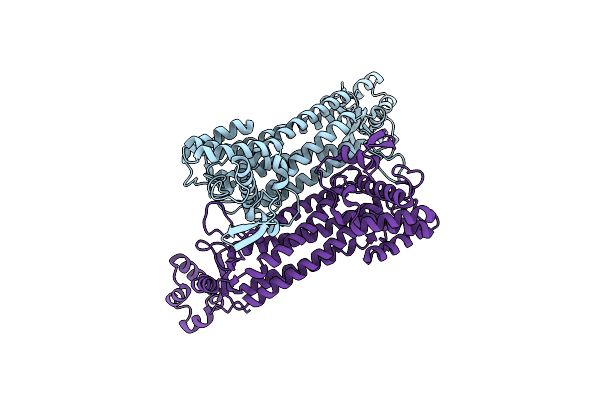 |
Crystal Structure Of Mutant Aspartase From Caenibacillus Caldisaponilyticus In The Closed Loop Conformation
Organism: Caenibacillus caldisaponilyticus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-01-15 Classification: LYASE |
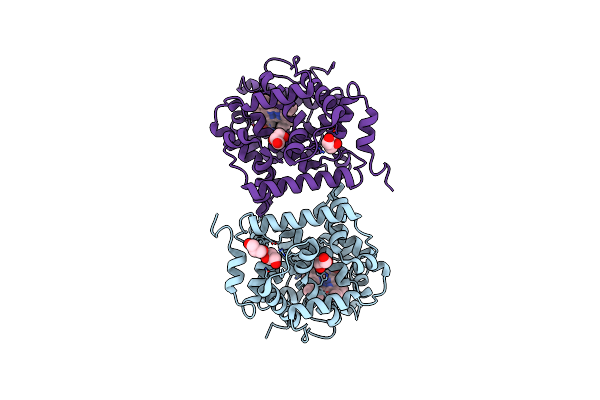 |
Crystal Structure Of T-Anethole Oxygenase From Stenotrophomonas Maltophilia
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, CL |
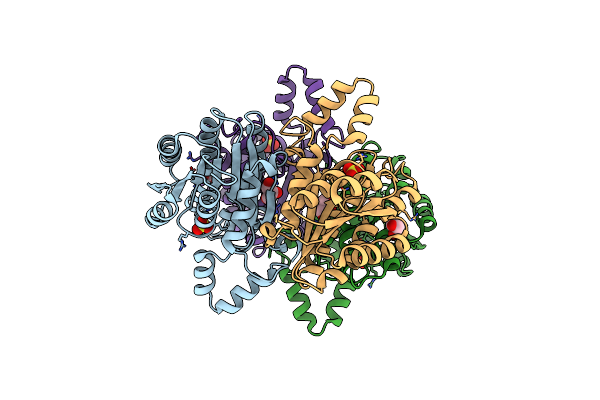 |
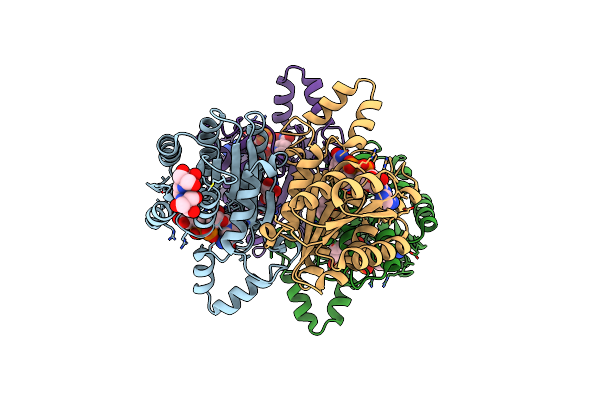
Crystal Structure Of Alcohol Dehydrogenase/Ketoreductase Variant From Thermus Thermophilus Apo Form
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
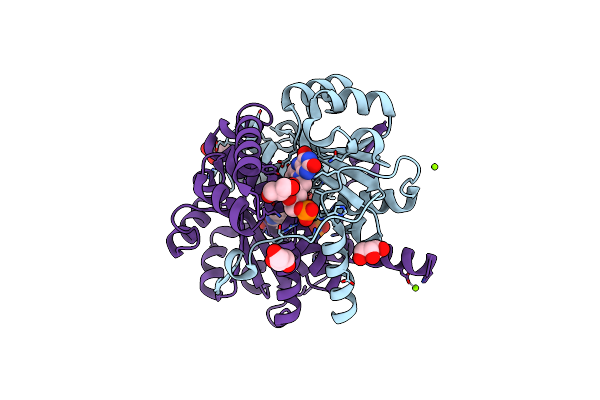
Crystal Structure Of Alcohol Dehydrogenase/Ketoreductase Variant From Thermus Thermophilus Apo Form
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: NAD, B3P, GOL |
 |
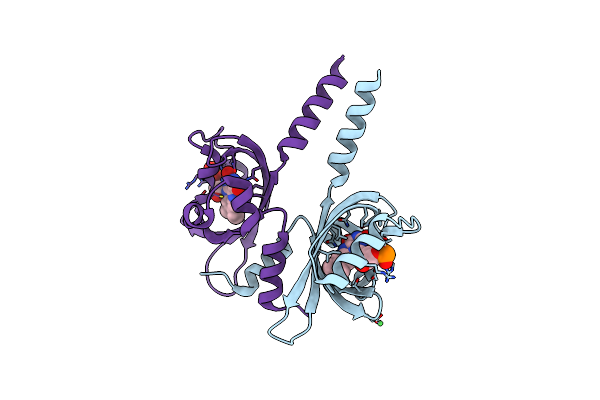
Organism: Bacillus tequilensis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, CL, MG |
 |
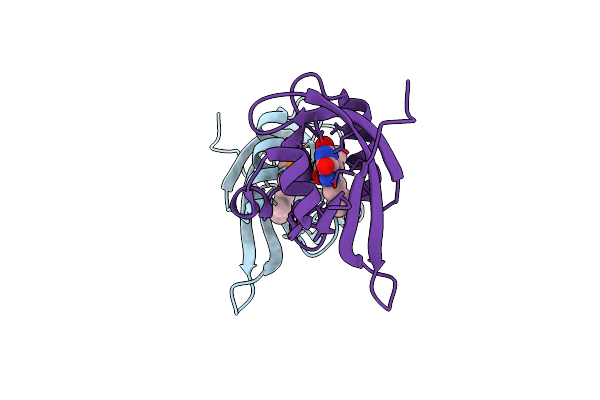
Crystal Structure Of Ppsb1-Lov Protein From Pseudomonas Putida With Covalent Fmn
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-11-29 Classification: SIGNALING PROTEIN Ligands: FMN, NI |
 |
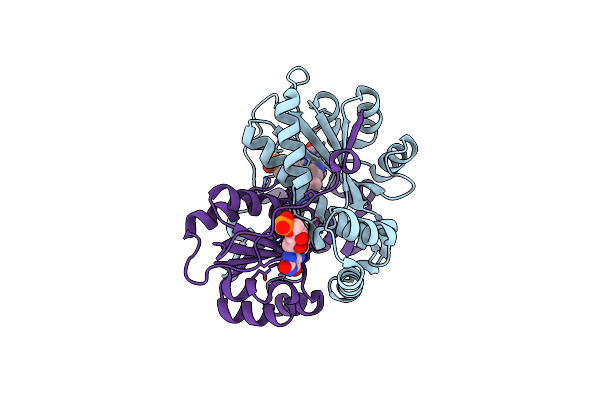
Crystal Structure Of Minisog Protein From Arabidopsis Thaliana With Covalent Fmn
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-29 Classification: SIGNALING PROTEIN Ligands: FMN |
 |
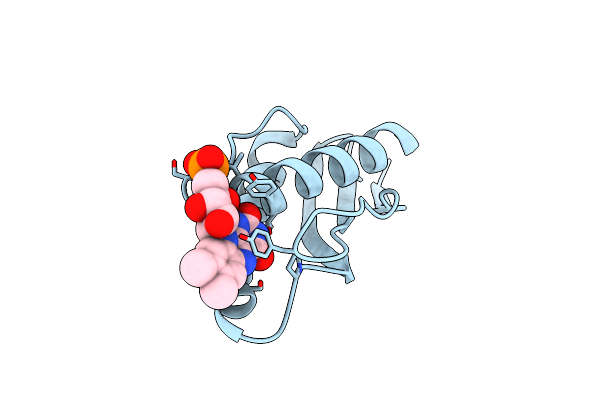
Crystal Structure Of Nitroreductase From Bacillus Tequilensis With Covalent Fmn
Organism: Bacillus tequilensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-29 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
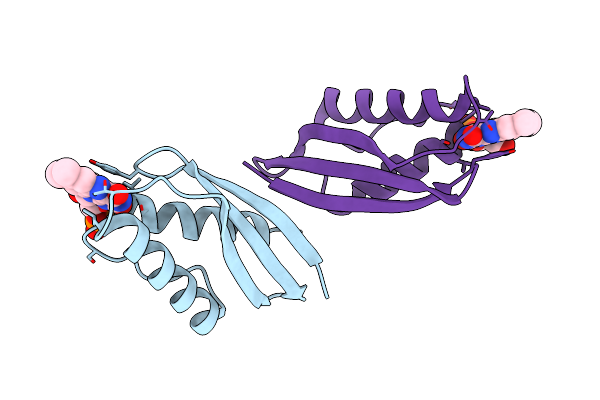
Organism: Streptomyces azureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-07-19 Classification: FLAVOPROTEIN Ligands: FMN |
 |
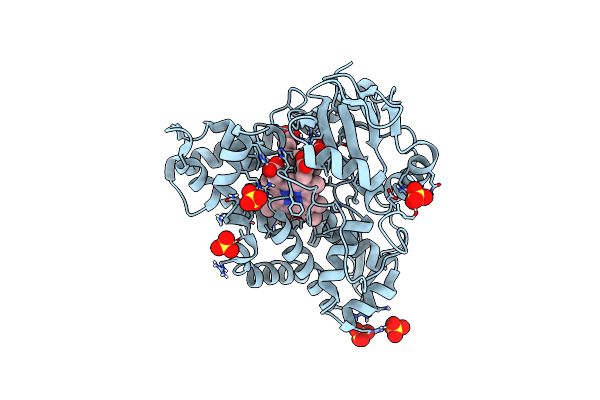
Organism: Clostridiaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-19 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Crystal Structure Of Cyp109A2 From Bacillus Megaterium Bound With Putative Ligands Hexanoic Acid And Octanoic Acid
Organism: Priestia megaterium dsm 319
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: HEB, 6NA, OCA, SO4 |
 |
Crystal Structure Of Cyp109A2 From Bacillus Megaterium Bound With Testosterone And Putative Ligand 4,6-Dimethyloctanoic Acid
Organism: Priestia megaterium dsm 319
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: HEB, W2L, TES, SO4 |
 |
Crystal Structure Of Holo-Swhpa-Mg (Hydroxy Ketone Aldolase) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate And D-Glyceraldehyde
Organism: Rhizorhabdus wittichii rw1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-23 Classification: LYASE Ligands: 3PY, 3GR, PEG, MG, BR, K |
 |
Crystal Structure Of S116A Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-11-16 Classification: LYASE Ligands: 3PY, MG |
 |
Crystal Structure Of Apo-Swhka (Hydroxy Ketone Aldolase) From Sphingomonas Wittichii Rw1
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-11-16 Classification: LYASE Ligands: PEG, BR, K |
 |
Crystal Structure Of Holo-H44A Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1, In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-16 Classification: LYASE Ligands: MG, BR, K, 3PY |
 |
Crystal Structure Of Holo-Swhpa-Mn (Hydroxyketoacid Aldolase) From Sphingomonas Wittichii Rw1
Organism: Rhizorhabdus wittichii rw1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-10-26 Classification: LYASE Ligands: MN, K, BR |
 |
Crystal Structure Of Holo-F210W Mutant Of Hydroxy Ketone Aldolase (Swhka)From Sphingomonas Wittichii Rw1
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-10-26 Classification: LYASE Ligands: PEG, MG, BR, K |
 |
Crystal Structure Of Holo-F210W Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-10-26 Classification: LYASE Ligands: 3PY, PEG, MG, K, BR |

