Search Count: 14
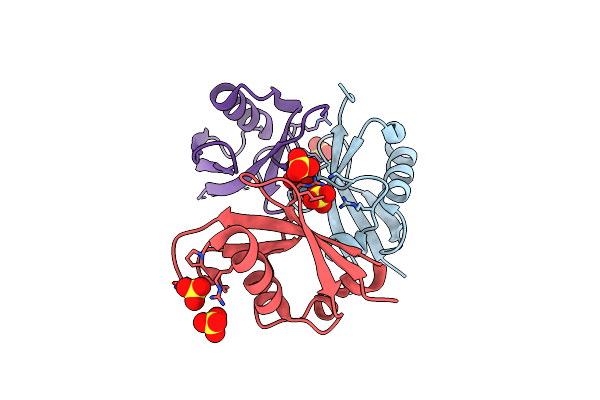 |
The Structure Of The Ubiquitin-Like Modifier Fat10 Reveals A Novel Targeting Mechanism For Degradation By The 26S Proteasome
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2018-08-29 Classification: SIGNALING PROTEIN Ligands: SO4 |
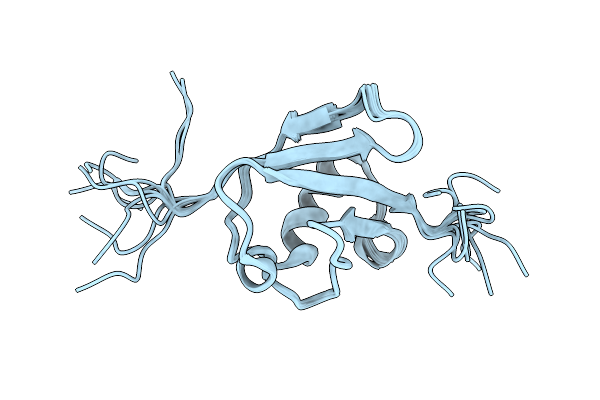 |
The Structure Of The Ubiquitin-Like Modifier Fat10 Reveals A Novel Targeting Mechanism For Degradation By The 26S Proteasome
|
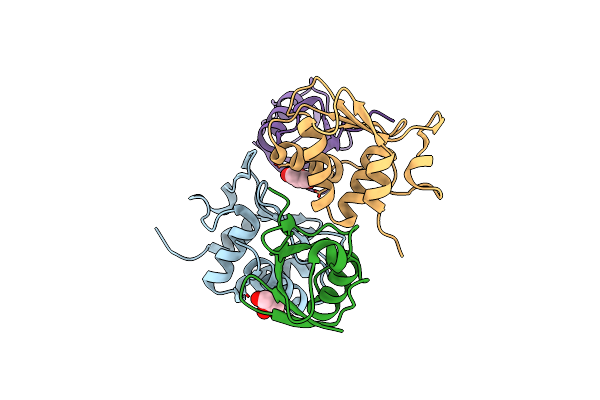 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-07-11 Classification: LIGASE Ligands: GOL |
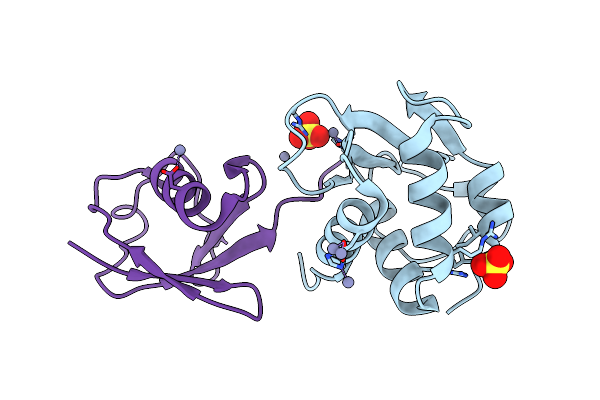 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2018-07-11 Classification: LIGASE Ligands: SO4, ZN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-02-21 Classification: PROTEIN BINDING Ligands: GOL |
 |
Crystal Structure Of An Arabinose Binding Protein With Designed Serotonin Binding Site In Open, Ligand-Free State
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-10-13 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2009-09-08 Classification: TRANSCRIPTION REGULATOR |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2008-08-19 Classification: SH3 DOMAIN/UBIQUITIN |
 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2007-02-13 Classification: SIGNALING PROTEIN, TRANSFERASE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-02-13 Classification: TRANSFERASE Ligands: MG, ADP |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2005-11-01 Classification: STRUCTURAL PROTEIN |
 |
 |
Solution Structure Of The Prp40 Ww Domain Pair Of The Yeast Splicing Factor Prp40
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2002-12-04 Classification: NUCLEAR PROTEIN |

