Search Count: 65
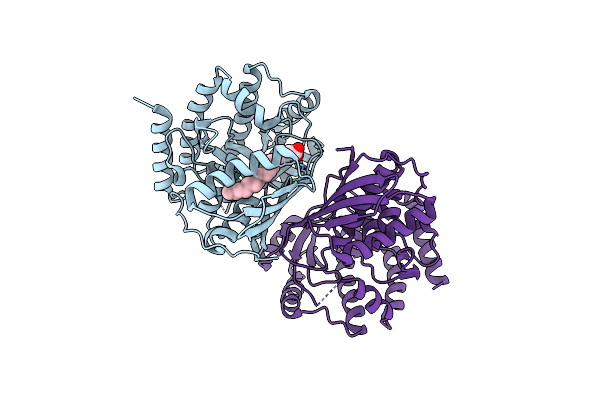 |
Crystal Structure Of Mevalonate 3,5-Bisphosphate Decarboxylase From Picrophilus Torridus
Organism: Picrophilus torridus dsm 9790
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-12-22 Classification: LYASE Ligands: OLA |
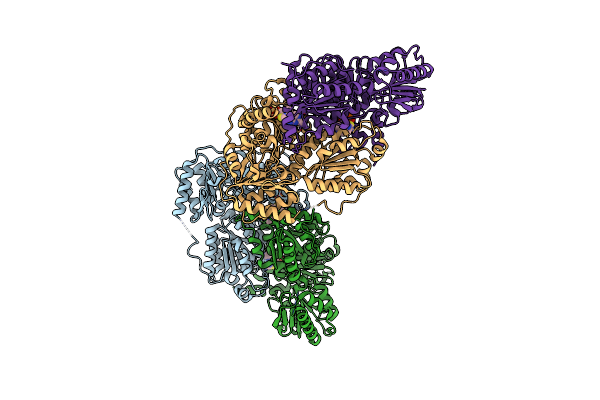 |
Alpha-Ketoisovalerate Decarboxylase (Kivd) From Lactococcus Lactis, Thermostable Mutant
Organism: Lactococcus lactis subsp. lactis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-08-05 Classification: LYASE Ligands: MG, TPP |
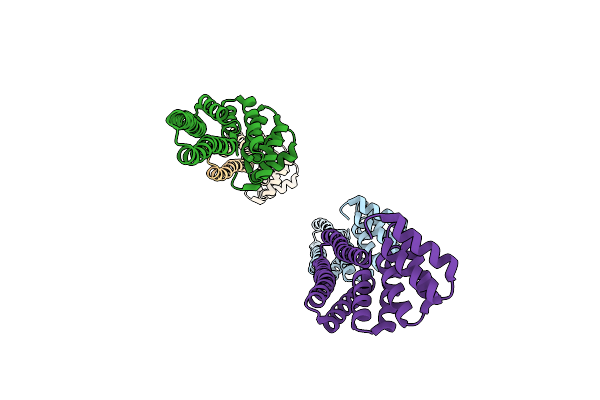 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.89 Å Release Date: 2018-03-14 Classification: MEMBRANE PROTEIN |
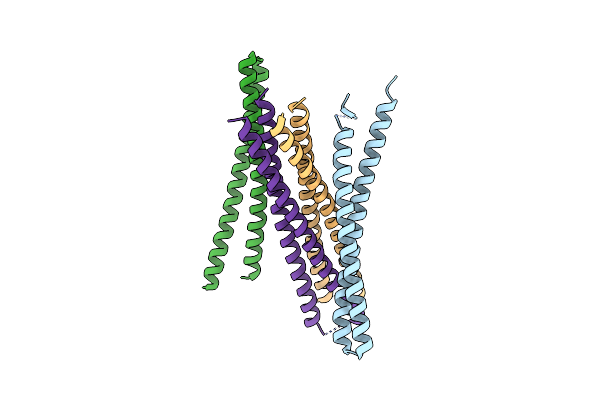 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2018-03-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Callithrix jacchus
Method: ELECTRON MICROSCOPY Release Date: 2017-10-11 Classification: TRANSPORT PROTEIN Ligands: Y01, NA, 3PE |
 |
Organism: Cyanothece sp. (strain pcc 8801)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-05-31 Classification: OXIDOREDUCTASE Ligands: FE, PGR, BU1, GOL, HEZ, POL, 1BO, PDO |
 |
Organism: Cyanothece sp. (strain pcc 8801)
Method: X-RAY DIFFRACTION Release Date: 2017-05-31 Classification: OXIDOREDUCTASE Ligands: FE, GOL, MES, IMD |
 |
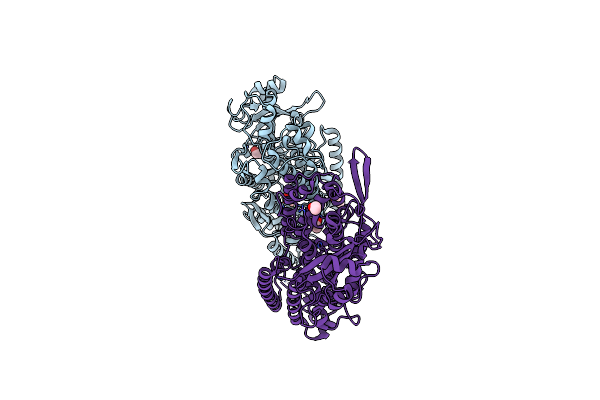
Organism: Cyanothece sp. (strain pcc 8801)
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2017-05-31 Classification: OXIDOREDUCTASE Ligands: FE, GOL, IMD, CL |
 |
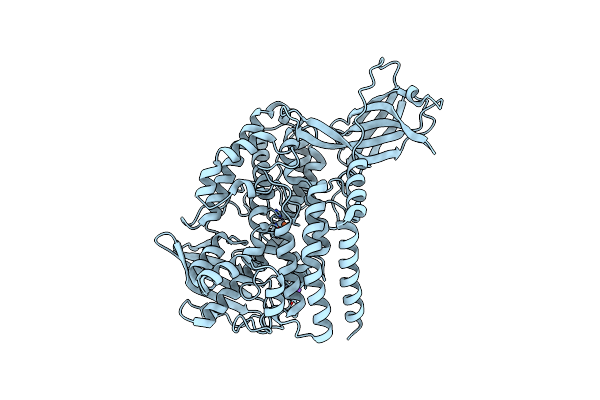
Organism: Cyanothece sp. (strain pcc 8801)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-05-31 Classification: OXIDOREDUCTASE Ligands: MN, CL, EOH |
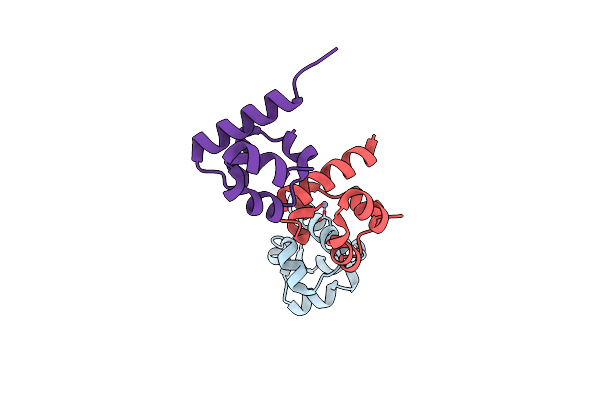 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-01-27 Classification: PROTEIN BINDING Ligands: ZN |
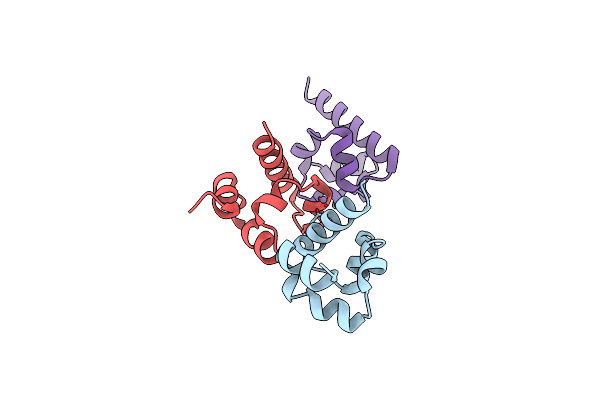 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-27 Classification: PROTEIN BINDING Ligands: ZN |
 |
Crystal Structure Of A 9R-Lipoxygenase From Cyanothece Pcc8801 At 2.7 Angstroms
Organism: Cyanothece sp. (strain pcc 8801)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-12-23 Classification: OXIDOREDUCTASE Ligands: FE2, NA |
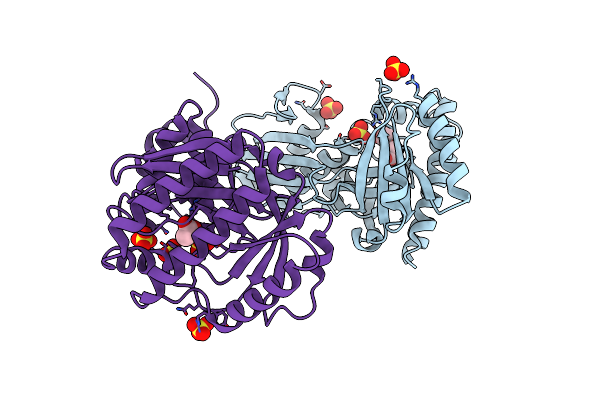 |
Crystal Structure Of Mevalonate-3-Kinase From Thermoplasma Acidophilum (Apo Form)
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: SO4, ACT |
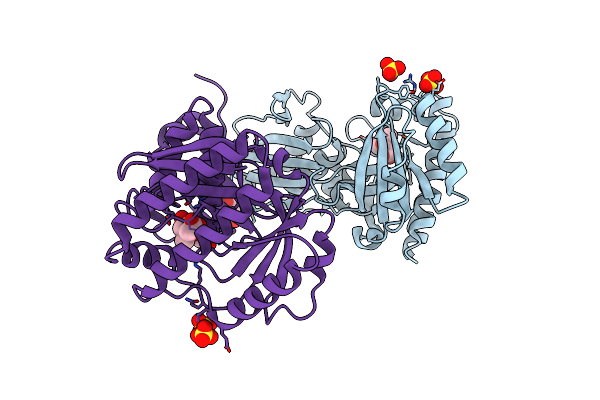 |
Crystal Structure Of Mevalonate-3-Kinase From Thermoplasma Acidophilum (Mevalonate Bound)
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: MEV, SO4, ACT, GOL |
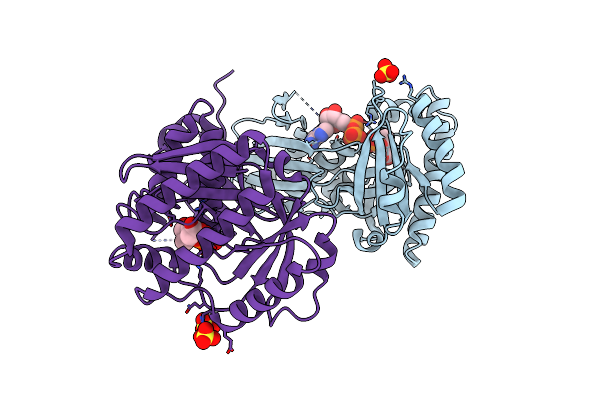 |
Crystal Structure Of Mevalonate-3-Kinase From Thermoplasma Acidophilum (Mevalonate 3-Phosphate/Adp Bound)
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: ADP, 3S4, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-07-16 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-07-16 Classification: STRUCTURAL PROTEIN Ligands: MG |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-05-22 Classification: HYDROLASE Ligands: CA, CL, GOL, 1PE, PE4 |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-02-06 Classification: HYDROLASE Ligands: CA, IPA, GOL, 1PE |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-02-06 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, DEP, PGE |

