Search Count: 35
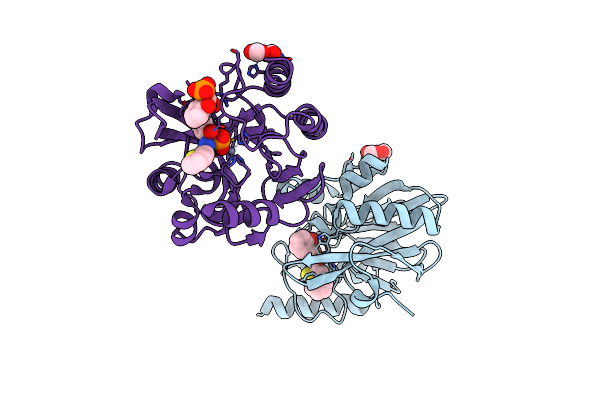 |
Vim-2 In Complex With Gkv61 (5C) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1IKA, ZN, A1H8T |
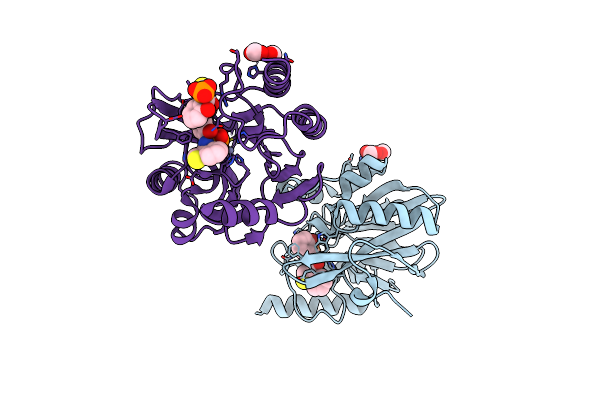 |
Vim-2 In Complex With Gkv53 (5D) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1IJ9, ZN, A1H8W |
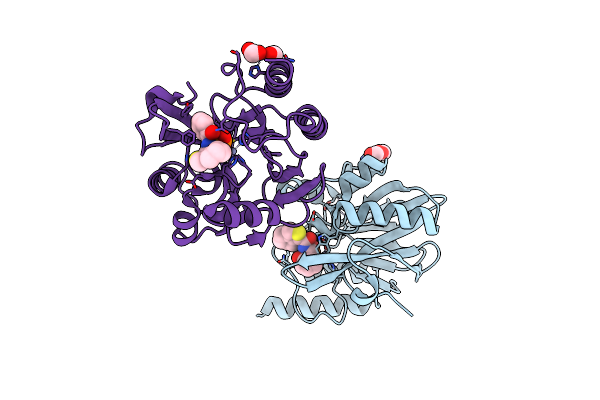 |
Vim-2 In Complex With Gkv65 (5G) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1H8V, ZN, MG |
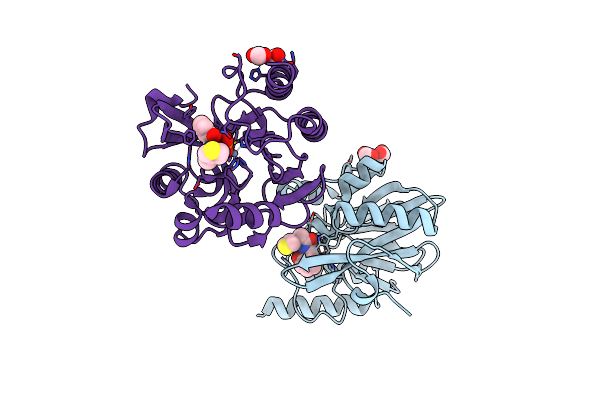 |
Vim-2 In Complex With Gkv63 (5J) - Dynamically Chiral Phosphonic Acid-Type Metallo-Beta-Lactamase Inhibitors
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-04-30 Classification: ANTIBIOTIC Ligands: FMT, A1H8U, ZN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: LYASE |
 |
Organism: Escherichia coli bl21
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: LYASE |
 |
Fructose-6-Phosphate Aldolase (Fsa) Mutant R134V, S166G, With Covalently Bound Active Site Ligand
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2023-02-08 Classification: LYASE |
 |
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant L259V Complexed With Fe, Nadh, And Glycerol
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-12-28 Classification: OXIDOREDUCTASE Ligands: NAI, GOL, FE |
 |
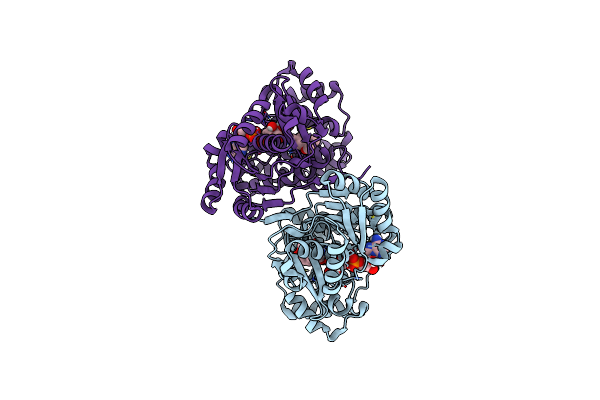
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant N151G, L259V Complexed With Fe, Nad, And Dimethoxyphenyl Acetamide
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-12-28 Classification: OXIDOREDUCTASE Ligands: E9I, APR, ACT, FE, GOL |
 |
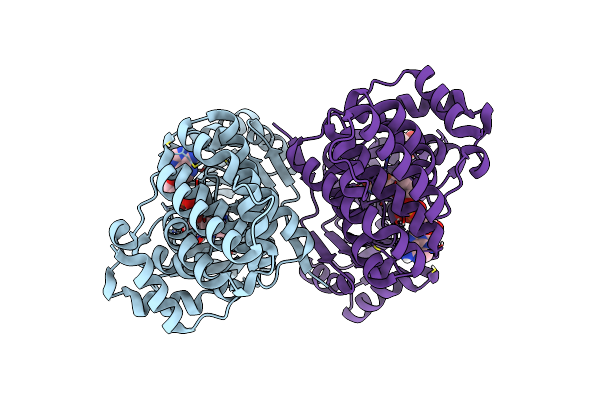
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant N151G, L259V Complexed With Fe, Nadh, And Dimethoxyphenyl Acetamide
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-12-28 Classification: OXIDOREDUCTASE Ligands: E9I, APR, FE |
 |
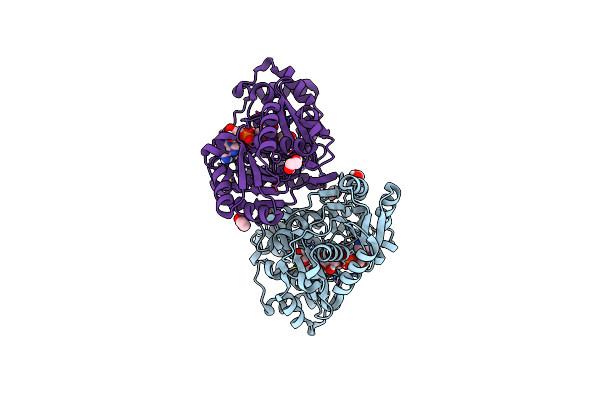
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant N151G, L259V Complexed With Fe, Nadh, And Glycerol
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-28 Classification: OXIDOREDUCTASE Ligands: NAI, FE, GOL |
 |
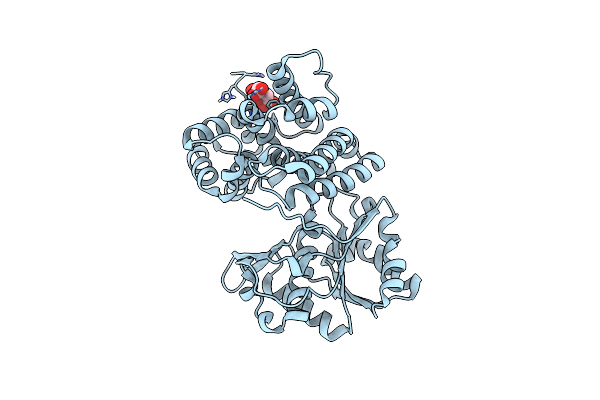
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant N151G, L259V Complexed With Fe, Nad+, And Ethylene Glycol
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: FE, EDO, FMT, APR |
 |
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: CIT |
 |
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant F254I Complexed With Fe, Nad+, And Glycerol
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: FE, GOL, NAD |
 |
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant F254I Complexed With Fe, Nad+, And Ethylene Glycol
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, FE |
 |
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant N151G, L259V Complexed With Fe, Nadh, And Glycerol (Absence Of Nicotinamide Ring)
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: APR, FE |
 |
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant F254I Complexed With Fe, Nadh, And Glycerol
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: NAI, APR, FE, GOL |
 |
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-05-25 Classification: HYDROLASE Ligands: OAA, DEP |
 |
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:0.87 Å Release Date: 2022-05-25 Classification: HYDROLASE Ligands: CIT, PO4, PPI |
 |
Organism: Rhodococcus sp. m8
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2018-12-19 Classification: OXIDOREDUCTASE Ligands: ZN, NAD |

