Search Count: 73
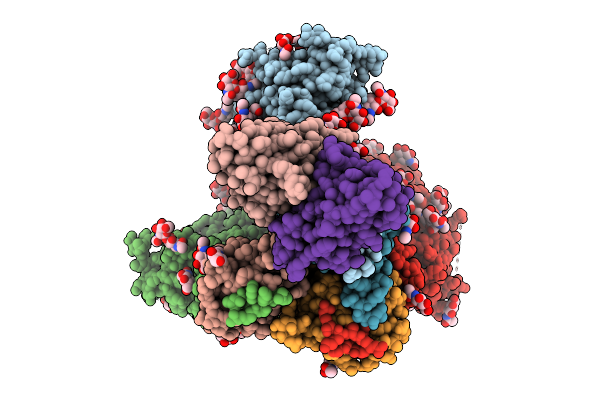 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN Ligands: NAG |
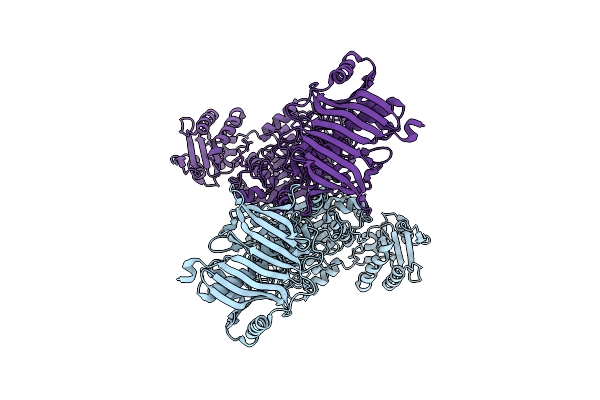 |
Organism: Mycetohabitans rhizoxinica
Method: ELECTRON MICROSCOPY Resolution:2.84 Å Release Date: 2024-04-03 Classification: TRANSFERASE |
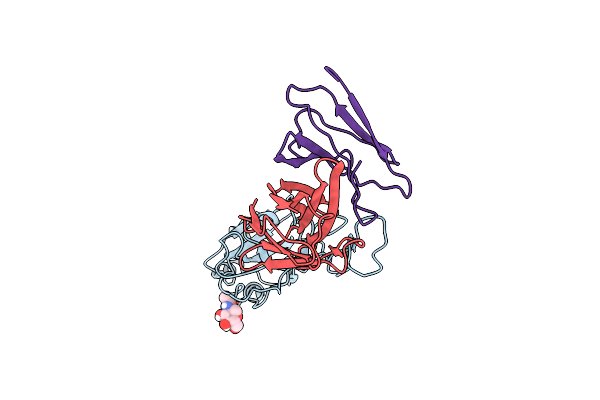 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: VIRAL PROTEIN Ligands: NAG |
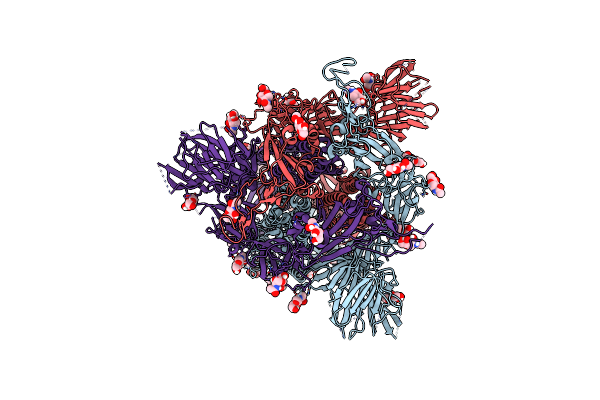 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Sars-Cov-2 S Omicron Spike B.1.1.529 - Rbd Up - 1-P2G3 And 1-P5C3 Fabs (Local)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2022-05-04 Classification: HYDROLASE Ligands: K36, B1S, EDO, CL, K, NA |
 |
Fgfr1 Kinase Domain (Residues 458-765) With Mutations C488A, C584S In Complex With 38.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-12-01 Classification: TRANSFERASE Ligands: 47I, SO4, EDO |
 |
Fgfr1 Kinase Domain (Residues 458-765) With Mutations C488A, C584S In Complex With 34.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-12-01 Classification: TRANSFERASE Ligands: 42I, SO4, EDO |
 |
Fgfr1 Kinase Domain (Residues 458-765) With Mutations C488A, C584S In Complex With 19.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-12-01 Classification: TRANSFERASE Ligands: 466, SO4, EDO, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-12-01 Classification: TRANSFERASE Ligands: 47I, SO4, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-10-13 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-10-13 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Bovine Cytochrome Bc1 In Complex With Tetrahydro-Quinolone Inhibitor Jag021
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-07-22 Classification: MEMBRANE PROTEIN Ligands: PG4, 6PE, PO4, CDL, HEM, LMT, PEE, JAG, HEC, FES, PX4 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-06-24 Classification: HYDROLASE Ligands: EDO, ZN, SO4, CL |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-06-24 Classification: HYDROLASE Ligands: KJK, MES, PEG, EDO, K9K, ZN, CL, NA |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2020-06-24 Classification: HYDROLASE Ligands: OK3, MPD, EPE, NA, CL, GOL |
 |
Crystal Structure Of Ampc From E. Coli With Cyclic Boronate 3 (Cb3 / Apc308)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-06-24 Classification: HYDROLASE Ligands: KL8, PEG, DMS, EDO, CL, ZN |
 |
Crystal Structure Of E. Coli Seryl-Trna Synthetase Complexed To Seryl Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-01-22 Classification: LIGASE Ligands: SSA, PO4, EDO |

