Search Count: 8
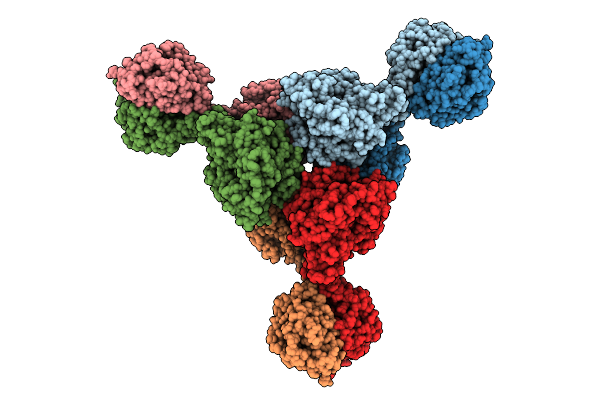 |
Crispr-Associated Deaminase Cad1 In Ca4 Bound In Hexamer Form Refined Against The Consensus Map
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: ANTIVIRAL PROTEIN/RNA Ligands: ZN |
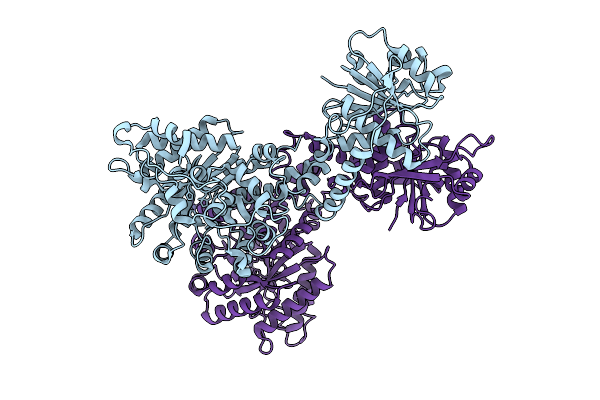 |
Cryoem Structure Of Cad1 In Apo Form, Symmetry Expanded Dimer, Refined Against A Composite Map
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ZN |
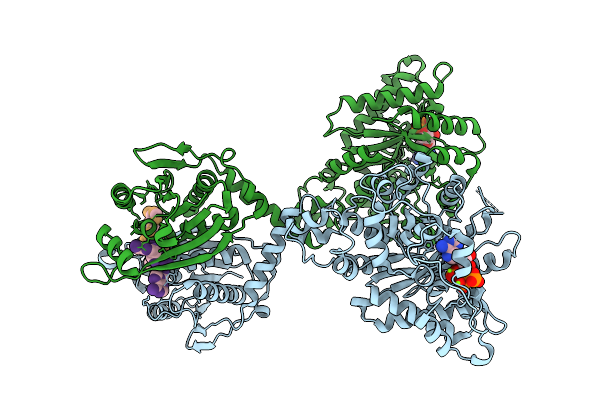 |
Cryoem Structure Of Cad1 Bound With Ca4 And Atp, Symmetry Expanded Dimer Refined Against A Composite Map
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ATP, ZN, MG |
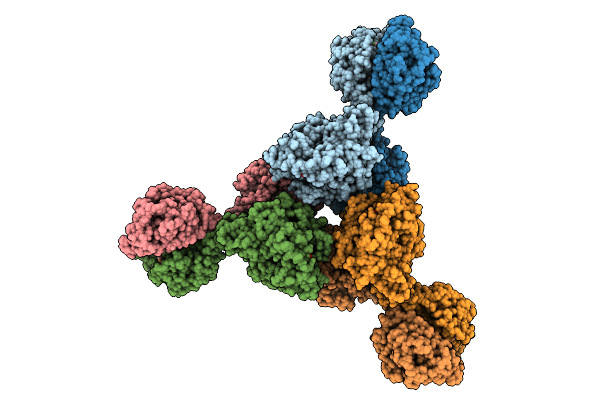 |
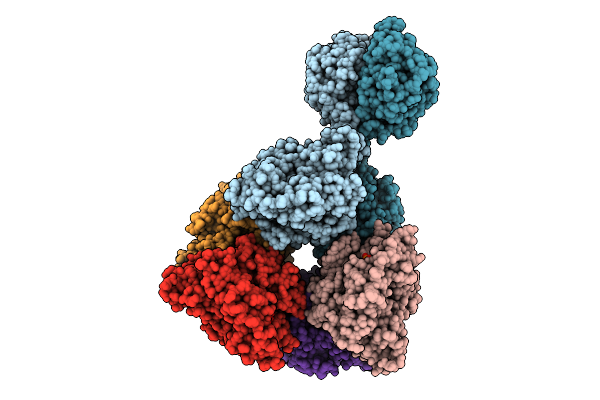
Cryoem Structure Of Cad1 Bound With Ca4 And Atp, Hexamer With Three Intact Dimers
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ATP, ZN, MG |
 |
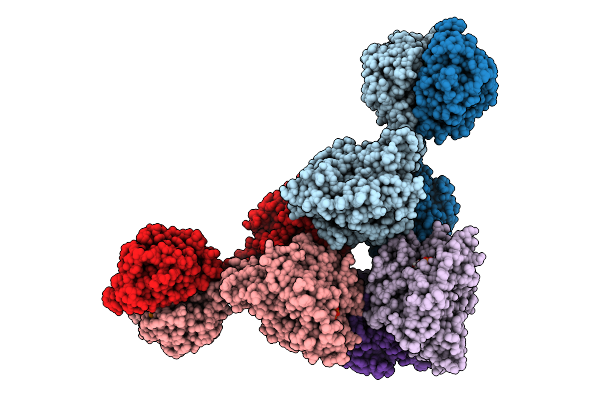
Cryoem Structure Of Cad1 Bound With Ca4 And Atp, Hexamer With One Intact Dimer
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ATP, ZN, MG |
 |
Cryoem Structure Of Cad1 Bound With Ca4 And Atp, Hexamer With Two Intact Dimers
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: IMMUNE SYSTEM, HYDROLASE Ligands: ATP, ZN, MG |
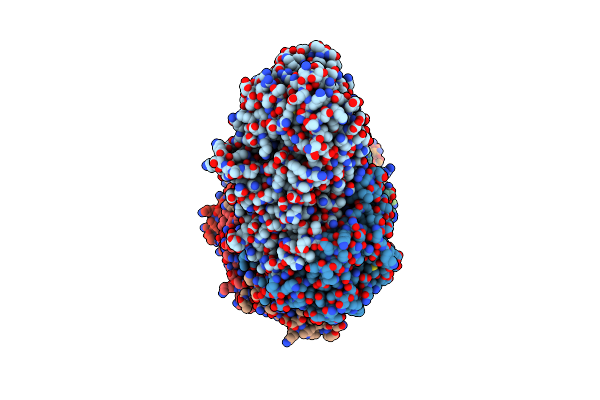 |
Cryo-Em Structure Of The Active Lactococcus Lactis Csm Bound To Target In Post-Cleavage Stage
Organism: Lactococcus lactis subsp. lactis
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: RNA BINDING PROTEIN/RNA Ligands: ATP, MG |
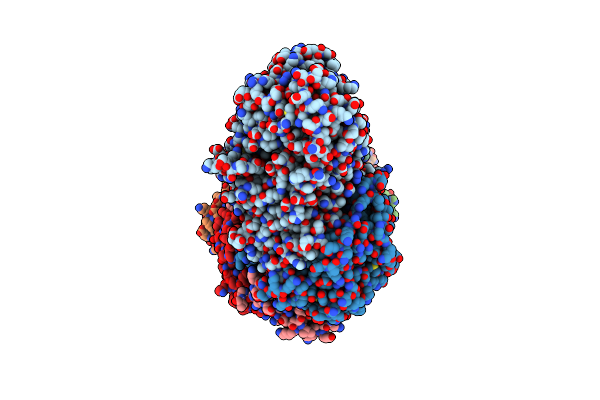 |
Cryo-Em Structure Of The Active Lactococcus Lactis Csm Bound To Target In Pre-Cleavage Stage
Organism: Lactococcus lactis subsp. lactis
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: RNA BINDING PROTEIN/RNA Ligands: ATP, MG, AMP |

