Search Count: 24
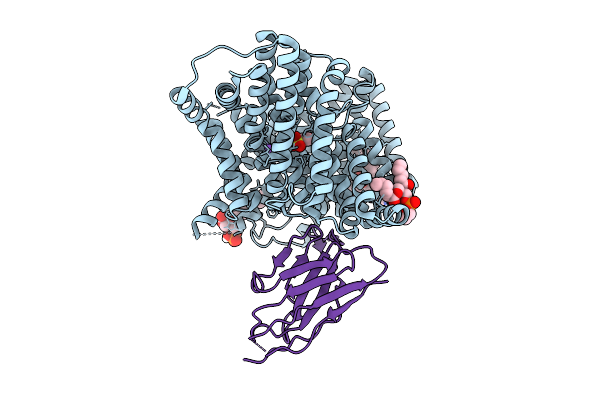 |
Cryo-Em Structure Of The Isethionate Trap Transporter Iseqm From Oleidesulfovibrio Alaskensis With Bound Isethionate
Organism: Oleidesulfovibrio alaskensis g20, Helicobacter pylori
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN |
 |
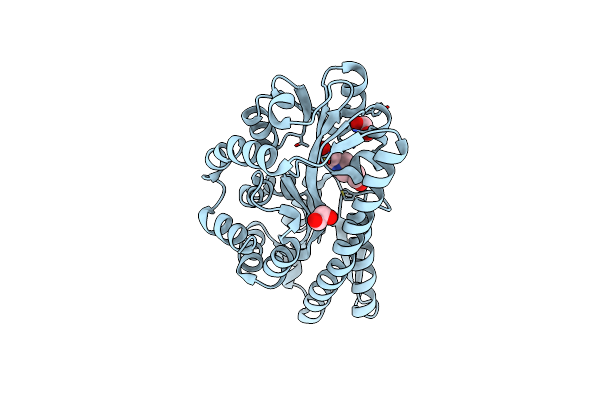
Crystal Structure Of A Mes Bound Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-08-28 Classification: TRANSPORT PROTEIN Ligands: MES |
 |
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-08-28 Classification: TRANSPORT PROTEIN Ligands: EPE, EDO |
 |
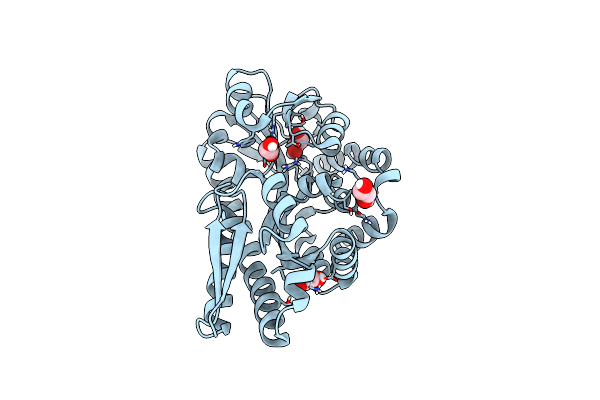
Crystal Structure Of An Isethionate Bound Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-07-10 Classification: TRANSPORT PROTEIN Ligands: 8X3, NO3, EDO |
 |
Apo Crystal Structure Of A Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-06-26 Classification: TRANSPORT PROTEIN Ligands: EDO, CL |
 |
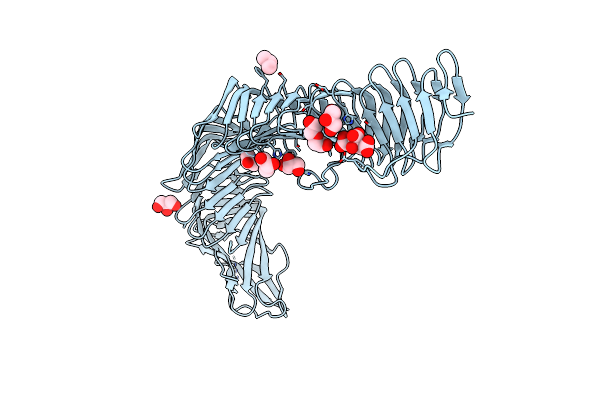
Structure Of Nad+ Glycohydrolase/Streptolysin O Complex From Group A Streptococcus
Organism: Streptococcus pyogenes a20
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-02-15 Classification: TOXIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2022-03-09 Classification: MICROBIAL PROTEIN Ligands: GOL, IPA, CIT, PEG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-03-09 Classification: MICROBIAL PROTEIN Ligands: GOL |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2022-03-09 Classification: MICROBIAL PROTEIN Ligands: PEG, P6G, GOL, MG, CL, SO4 |
 |
Structure Of Caulobacter Crescentus Suppressor Of Copper Sensitivity Protein C
Organism: Caulobacter vibrioides (strain atcc 19089 / cb15)
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2022-03-02 Classification: OXIDOREDUCTASE |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2021-04-21 Classification: OXIDOREDUCTASE Ligands: MG, EDO, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-02-05 Classification: NUCLEAR PROTEIN Ligands: ZN |
 |
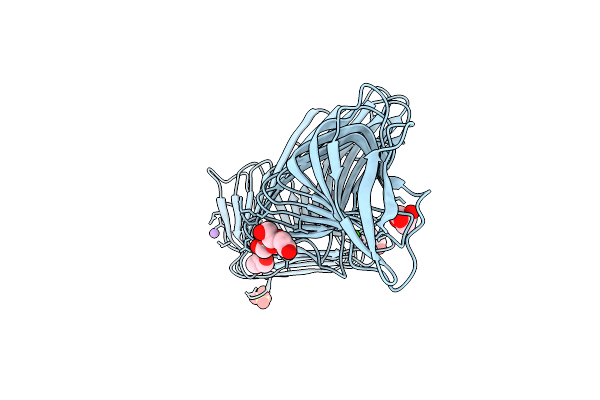
Crystal Structure Of The Thiol-Disulfide Exchange Protein Alpha-Dsba2 From Wolbachia Pipientis
Organism: Wolbachia endosymbiont of drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-04-17 Classification: ISOMERASE |
 |
Organism: Escherichia coli o6:h1
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-04-10 Classification: PROTEIN BINDING Ligands: GOL, P6G, SO4, CA, CL, NA |
 |
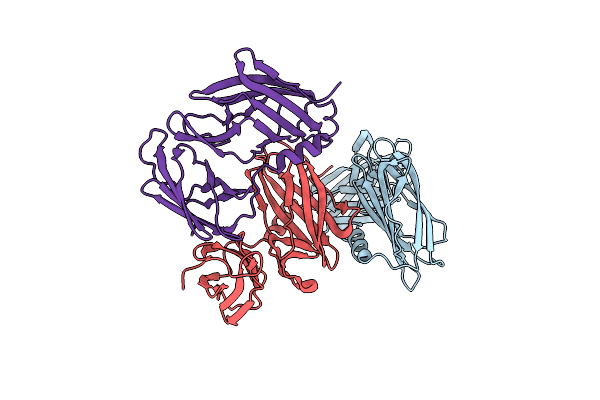
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-03-06 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of The N-Terminal Periplasmic Domain Of Scsb From Proteus Mirabilis
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-03-07 Classification: OXIDOREDUCTASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2017-08-02 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2017-07-26 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-06-08 Classification: ISOMERASE |

