Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
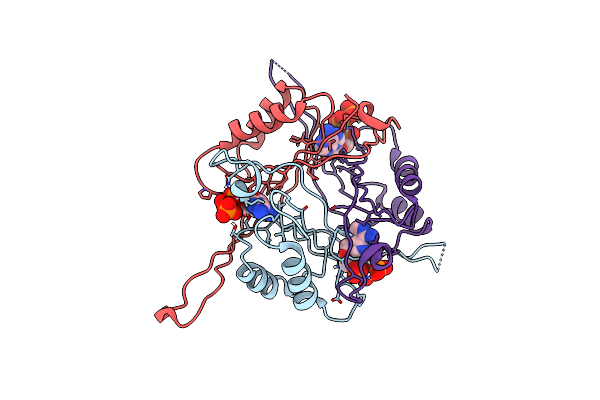 |
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-12-30 Classification: TRANSCRIPTION Ligands: ATP |
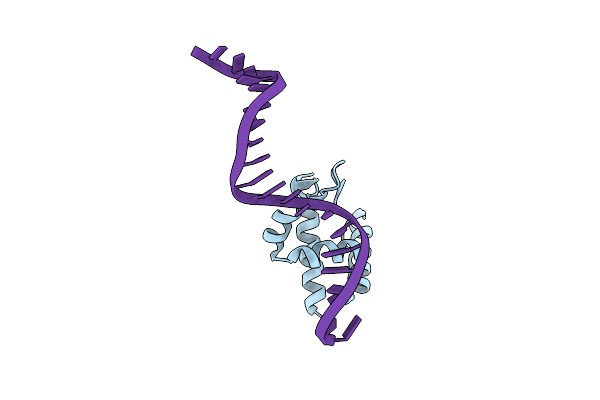 |
Organism: Bacillus megaterium, Synthetic dna
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-03-04 Classification: transcription/DNA |
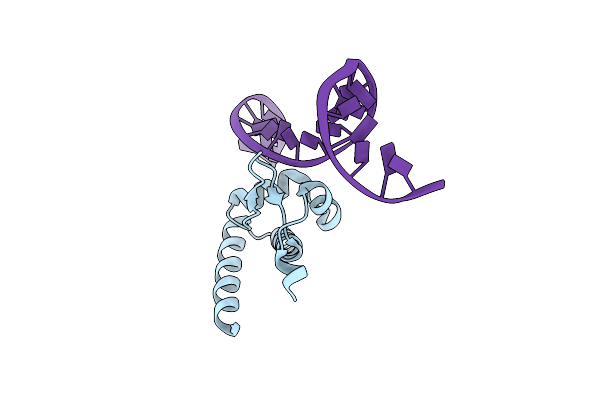 |
Organism: Bacillus megaterium, Synthetic dna
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-03-04 Classification: transcrption/DNA |
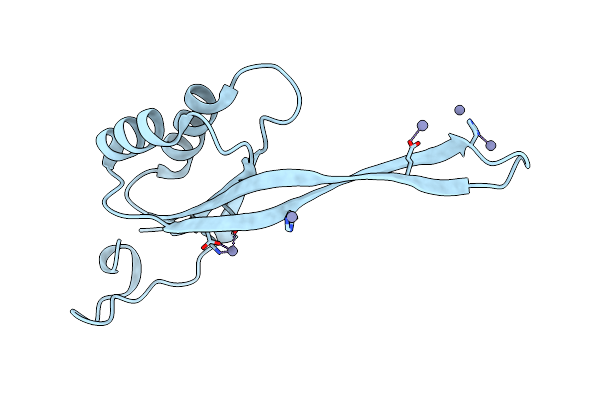 |
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2015-03-04 Classification: TRANSCRIPTION Ligands: ZN |
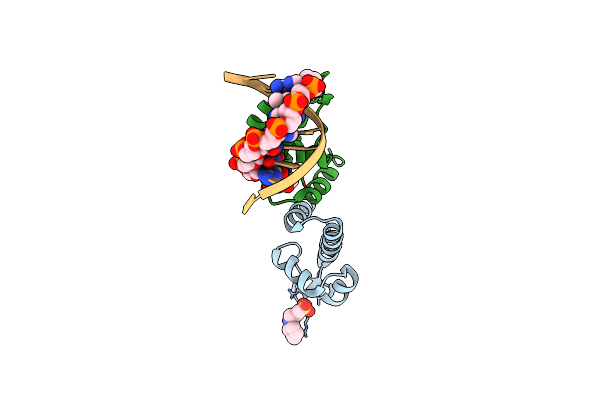 |
Organism: Bacillus subtilis subsp. subtilis, Synthetic dna
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2015-03-04 Classification: Transcription regulator/DNA Ligands: CXS |
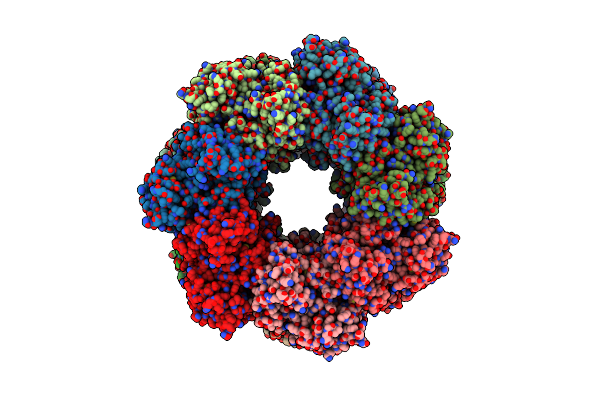 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2015-03-04 Classification: Ligase, chaperone Ligands: GLN, MG |
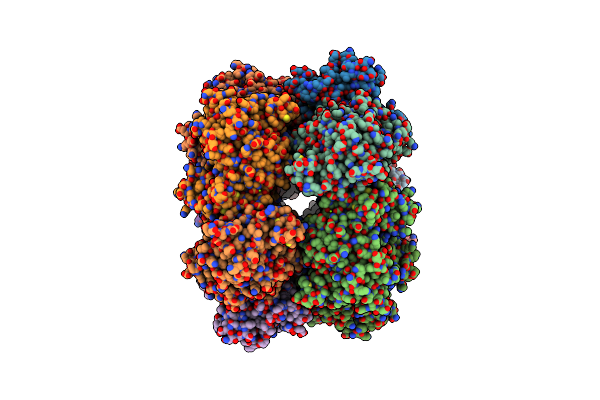 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Structure Of Gs-Q
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2013-11-13 Classification: LIGASE Ligands: MG, GLN, PO4 |
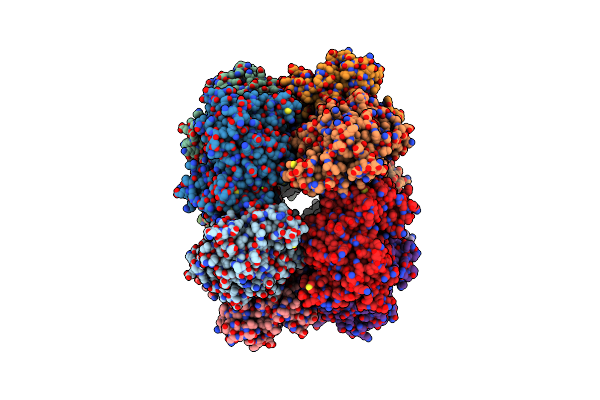 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Structure Of Apo Form Of Gs
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-11-13 Classification: LIGASE Ligands: MG, SO4 |
 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Form Two Of Gs-1
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2013-11-13 Classification: LIGASE Ligands: MG, GLN |
 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Structure Of The Transition State Complex
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2013-11-06 Classification: LIGASE Ligands: P3S, ADP, MG |
 |
B. Subtilis Glutamine Synthetase Structures Reveal Large Active Site Conformational Changes And Basis For Isoenzyme Specific Regulation: Structure Of Gs-Glutamate-Amppcp Complex
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2013-10-30 Classification: LIGASE Ligands: GLU, ADP, MG, SO4 |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-06-19 Classification: dna binding protein/DNA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2013-06-19 Classification: dna binding protein/DNA Ligands: MES |
 |
Organism: Vibrio cholerae o1 biovar el tor
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2013-06-19 Classification: dna binding protein/DNA |
 |
Organism: Vibrio cholerae o1 biovar el tor
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-06-19 Classification: DNA BINDING PROTEIN |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-06-19 Classification: DNA BINDING PROTEIN |
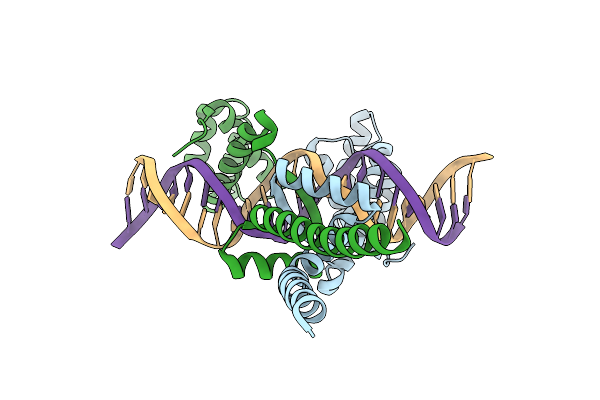 |
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2012-11-21 Classification: DNA BINDING PROTEIN/DNA |
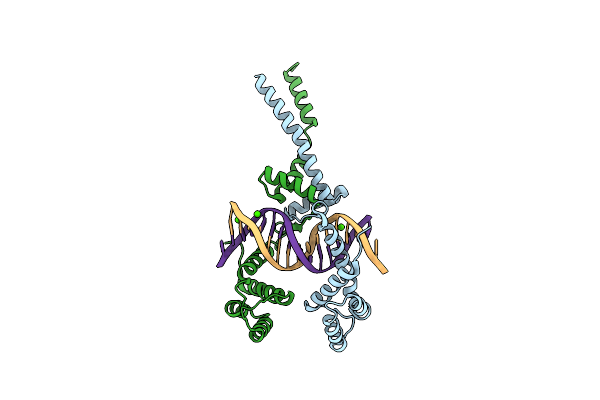 |
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-11-21 Classification: DNA BINDING PROTEIN/DNA Ligands: CA |
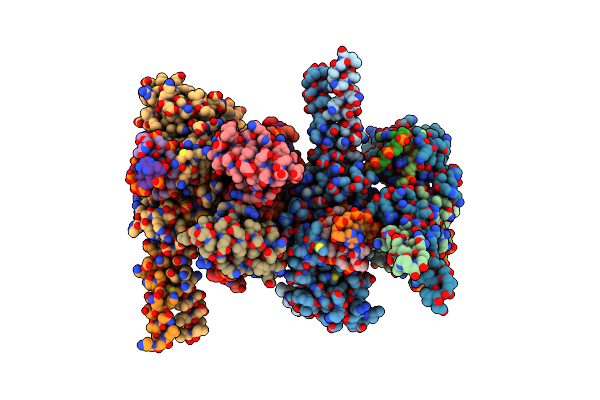 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2012-11-21 Classification: DNA BINDING PROTEIN |