Search Count: 35
 |
Panoptes Opts Minimal Crispr Polymerase (Mcpol) From Klebsiella Pneumoniae Strain Kp67
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ANTIVIRAL PROTEIN |
 |
Panoptes Opts Minimal Crispr Polymerase (Mcpol) With Non-Hydrolyzable Ligand Apcpp From Klebsiella Pneumoniae Strain Kp67
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ANTIVIRAL PROTEIN Ligands: APC, MG |
 |
Organism: Ensifer aridi
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2024-06-12 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
Organism: Ensifer aridi
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-06-12 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
Organism: Ensifer aridi
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2024-06-12 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
Organism: Ensifer aridi
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2024-06-12 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
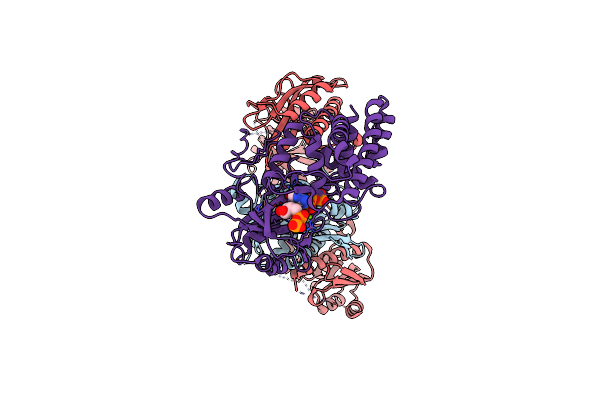
Organism: Enterobacter cloacae
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: TRANSFERASE Ligands: AMP, MG, ATP, ADP |
 |
Organism: Enterobacter cloacae
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: TRANSFERASE Ligands: AMP, MG, ATP, ADP |
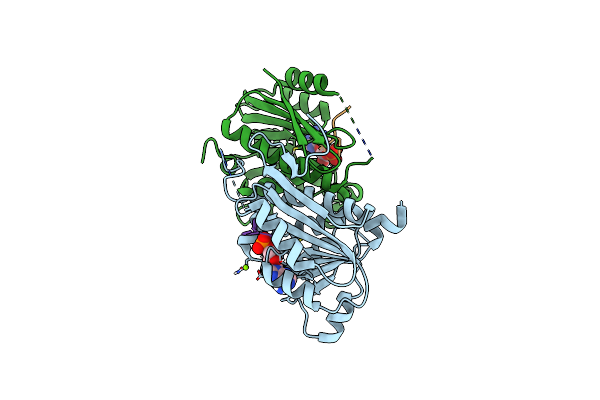 |
Structure Of Enterobacter Cloacae Cap2 Bound To Cdnd02 C-Terminus, Amp State
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-01-11 Classification: TRANSFERASE Ligands: MG, AMP |
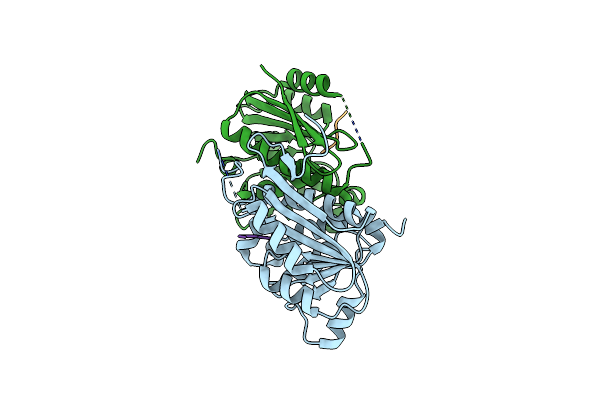 |
Structure Of Enterobacter Cloacae Cap2 Bound To Cdnd02 C-Terminus, Apo State
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-01-11 Classification: TRANSFERASE |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: MG, ATP, EDO |
 |
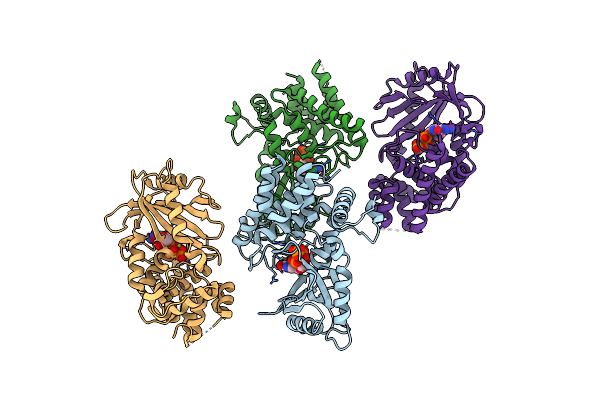
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: MG, GTP |
 |
Structure Of The Bradyrhizobium Diazoefficiens Cd-Ntase Cdng In Complex With Gtp
Organism: Bradyrhizobium diazoefficiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: GTP, MG |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: ADP, MG |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2020-09-09 Classification: CYTOSOLIC PROTEIN Ligands: CL |
 |
Crystal Structure Of Listeria Monocytogenes Cbpb Protein (Lmo1009) In Complex With C-Di-Amp
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-09-09 Classification: CYTOSOLIC PROTEIN Ligands: 2BA |
 |
Structure Of Moraxella Osloensis Cap4 Saved/Carf-Domain Containing Receptor
Organism: Moraxella osloensis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-06-17 Classification: HYDROLASE Ligands: MG |
 |
Structure Of Acinetobacter Baumannii Cap4 Saved/Carf-Domain Containing Receptor With The Cyclic Trinucleotide 2'3'3'-Caaa
Organism: Acinetobacter sp. atcc 27244, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-06-17 Classification: HYDROLASE Ligands: SO4 |
 |
Structure Of Acinetobacter Baumannii Cap4 Saved/Carf-Domain Containing Receptor
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-06-17 Classification: HYDROLASE Ligands: SO4 |
 |
Structure Of Acinetobacter Baumannii Cap4 Saved/Carf-Domain Containing Receptor With The Cyclic Trinucleotide 3'3'3'-Caaa
Organism: Acinetobacter baumannii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-06-17 Classification: HYDROLASE Ligands: SO4 |

