Search Count: 158
 |
Organism: Paenibacillus popilliae
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TOXIN Ligands: MG |
 |
Organism: Paenibacillus popilliae
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TOXIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: HYDROLASE Ligands: MG, ATP, ADP |
 |
Lysozyme Structure Based On Automated Real-Time Serial Crystallography Data Processing Using Crystfel
Organism: Gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: CL, NA |
 |
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2023-12-06 Classification: OXIDOREDUCTASE |
 |
Serial Crystallography Structure Of Cofactor-Free Urate Oxidase In Complex With The 5-Peroxo Derivative Of 9-Methyl Uric Acid At Room Temperature
Organism: Aspergillus flavus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-04-26 Classification: OXIDOREDUCTASE Ligands: XDS |
 |
Organism: Adeno-associated virus - 4
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: VIRUS LIKE PARTICLE Ligands: MG |
 |
Organism: Adeno-associated virus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: VIRUS LIKE PARTICLE |
 |
Adeno-Associated Virus Go.1 In Complex With Its Cellular Receptor Aavr At 2.4 Angstroms Resolution, Aavgo.1 Aavr
Organism: Homo sapiens, Adeno-associated virus
Method: ELECTRON MICROSCOPY Resolution:2.40 Å Release Date: 2022-11-23 Classification: VIRUS LIKE PARTICLE |
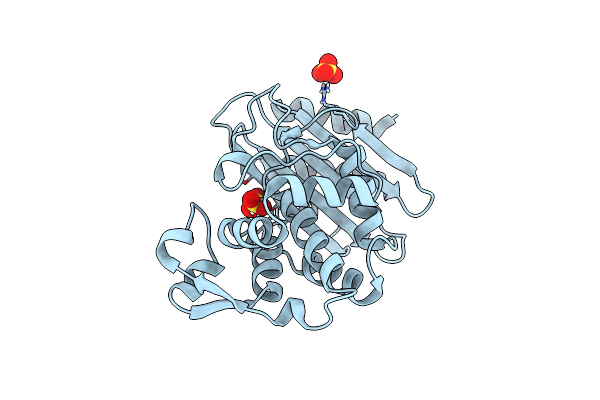 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-11-16 Classification: HYDROLASE Ligands: SO4 |
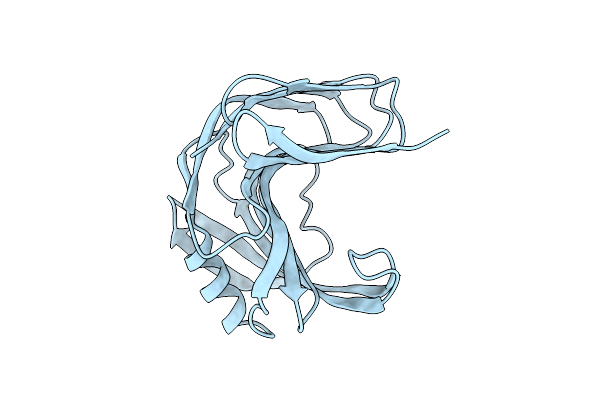 |
Room Temperature Ssx Structure Of Gh11 Xylanase From Nectria Haematococca (1000 Frames)
Organism: Fusarium haematococcum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-16 Classification: HYDROLASE |
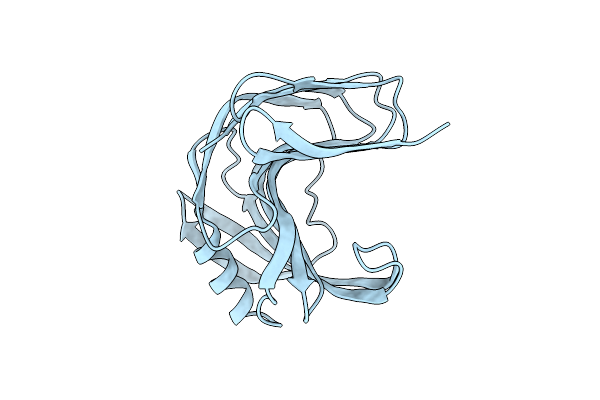 |
Room Temperature Ssx Structure Of Gh11 Xylanase From Nectria Haematococca (40000 Frames)
Organism: Fusarium haematococcum
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-11-16 Classification: HYDROLASE |
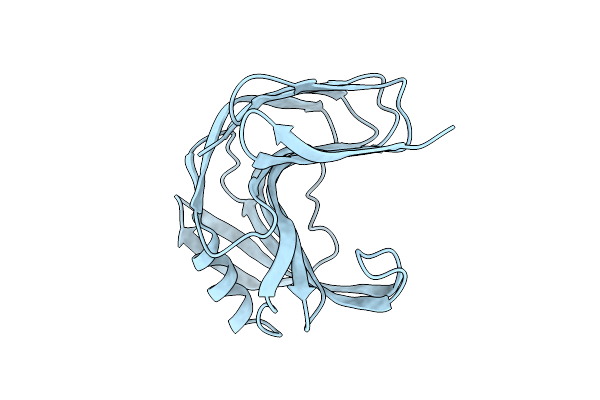 |
Room Temperature Ssx Structure Of Gh11 Xylanase From Nectria Haematococca (10000 Frames)
Organism: Fusarium haematococcum
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-11-16 Classification: HYDROLASE |
 |
Room Temperature Ssx Structure Of Gh11 Xylanase From Nectria Haematococca (4000 Frames)
Organism: Fusarium haematococcum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-11-16 Classification: HYDROLASE |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-11-16 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-11-16 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MEMBRANE PROTEIN |
 |
Structure Of Bam In Msp1E3D1 Nanodiscs Prepared From E. Coli Outer Membranes
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: MEMBRANE PROTEIN |

