Search Count: 412
 |
Organism: Enterobacteria phage cus-3
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: VIRUS |
 |
Heligmosomoides Polygyrus Tgf-Beta Mimic 6 Domain 3 (Tgm6-D3) Bound To Human Tgf-Beta Type Ii Receptor Extracellular Domain
Organism: Homo sapiens, Heligmosomoides polygyrus
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: IMMUNOSUPPRESSANT Ligands: GOL, CL, NA, CAC |
 |
Organism: Mycobacterium phage bxb1
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: VIRAL PROTEIN |
 |
Organism: Mycobacterium phage bxb1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: VIRAL PROTEIN |
 |
Mycobacteriophage Bxb1 Portal And Connector Assembly - Composite Map And Model
Organism: Mycobacterium phage bxb1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: VIRAL PROTEIN |
 |
Organism: Mycobacterium phage bxb1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: VIRAL PROTEIN |
 |
Organism: Mycobacterium phage bxb1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: VIRAL PROTEIN |
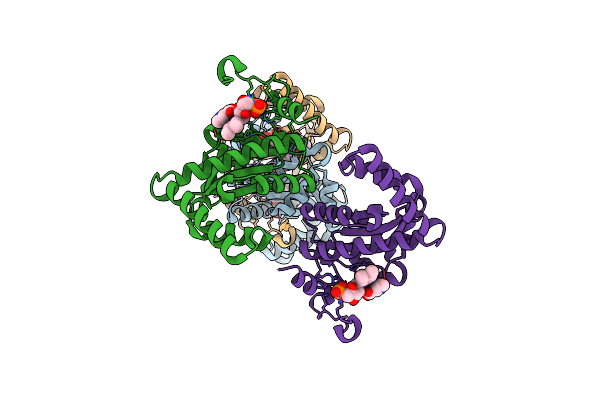 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: FMN, NIO |
 |
E. Coli Nfsb With The Unnatural Amino Acid P-Nitrophenylalanine At Position 124.
Organism: Escherichia coli dh5[alpha]
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: FMN, ACT |
 |
Organism: Mycobacterium phage adjutor
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: VIRUS |
 |
In Situ Cryo-Em Structure Of Bacteriophage P22 Tail Hub Protein: Tailspike Protein Complex At 2.8A Resolution
Organism: Salmonella phage p22
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: VIRAL PROTEIN |
 |
In Situ Cryo-Em Structure Of Bacteriophage P22 Portal Protein: Head-To-Tail Protein Complex At 3.0A Resolution
Organism: Salmonella phage p22
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: VIRAL PROTEIN |
 |
In Situ Cryo-Em Structure Of Bacteriophage P22 Tailspike Protein Complex At 3.4A Resolution
Organism: Salmonella phage p22
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: VIRAL PROTEIN |
 |
In Situ Cryo-Em Structure Of Bacteriophage P22 Gp1:Gp4:Gp5:Gp10:Gp9 N-Term Complex In Conformation 1 At 3.2A Resolution
Organism: Salmonella phage p22
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: VIRAL PROTEIN |
 |
In Situ Cryo-Em Structure Of Bacteriophage P22 Gp1:Gp5:Gp4: Gp10: Gp9 N-Term Complex In Conformation 2 At 3.1A Resolution
Organism: Salmonella phage p22
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: VIRAL PROTEIN |
 |
Organism: Tequatrovirus t4, Tequatrovirus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:4.98 Å Release Date: 2023-06-28 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
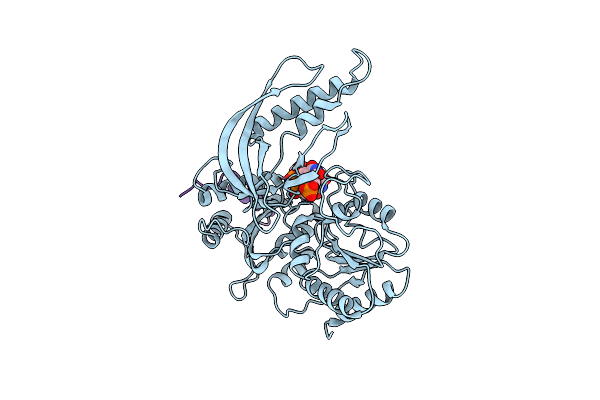 |
Organism: Tequatrovirus, Tequatrovirus t4, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.53 Å Release Date: 2023-06-28 Classification: DNA BINDING PROTEIN |
 |
Organism: Mycobacterium phage patience
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: VIRUS |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-04-19 Classification: OXIDOREDUCTASE Ligands: FMN, NIO, EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-04-19 Classification: OXIDOREDUCTASE Ligands: FMN, EDO, ACT |

