Search Count: 26
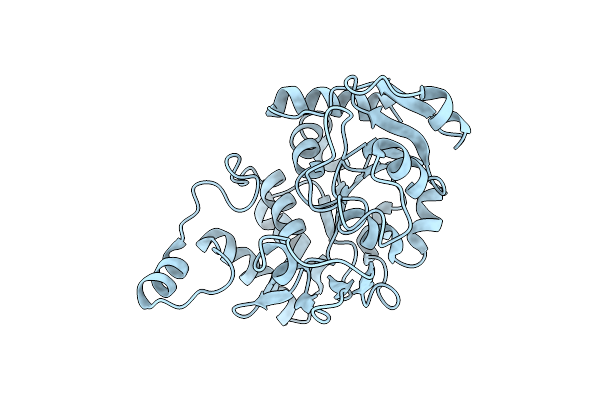 |
Crystal Structure Of 2-Phospho-(S)-Lactate Transferase From Methanosarcina Mazei. Northeast Structural Genomics Consortium Target Mar46
Organism: Methanosarcina mazei go1
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2008-03-18 Classification: TRANSFERASE |
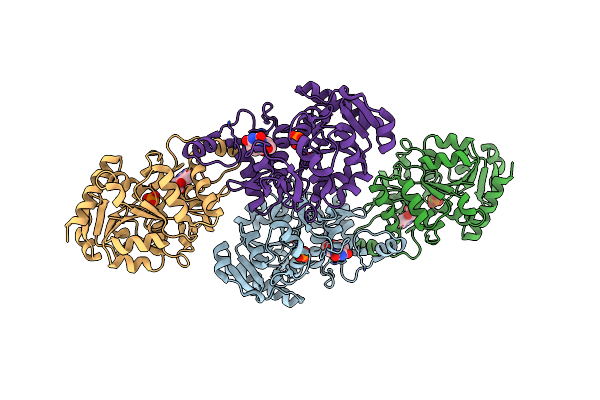 |
Crystal Structure Of 2-Phospho-(S)-Lactate Transferase From Methanosarcina Mazei In Complex With Fo And Phosphate. Northeast Structural Genomics Consortium Target Mar46
Organism: Methanosarcina mazei go1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-02-19 Classification: TRANSFERASE Ligands: PO4, FO1 |
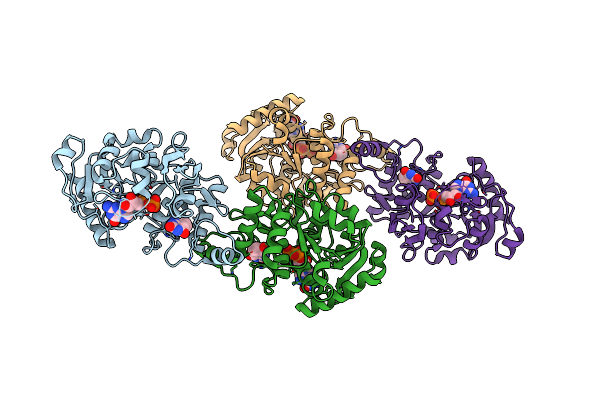 |
Crystal Structure Of 2-Phospho-(S)-Lactate Transferase From Methanosarcina Mazei In Complex With Fo And Gdp. Northeast Structural Genomics Consortium Target Mar46
Organism: Methanosarcina mazei go1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2008-02-19 Classification: TRANSFERASE Ligands: FO1, GDP |
 |
Crystal Structure Of Faicar Synthetase (Purp) From M. Jannaschii Complexed With Amppcp And Aicar
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-12-04 Classification: LIGASE Ligands: SO4, CL, ACP, AMZ |
 |
Crystal Structure Of Faicar Synthetase (Purp) From M. Jannaschii Complexed With Atp And Aicar
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-12-04 Classification: LIGASE Ligands: SO4, CL, ATP, AMZ |
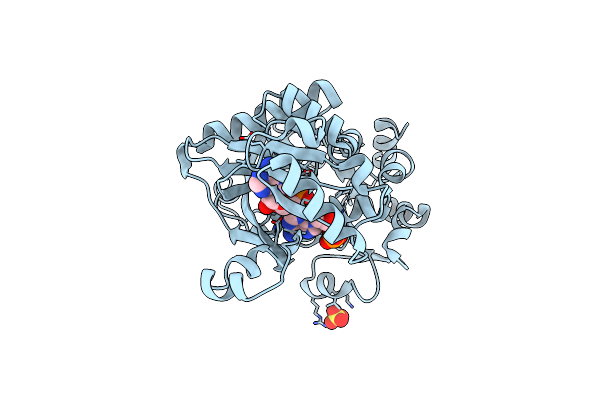 |
Crystal Structure Of Faicar Synthetase (Purp) From M. Jannaschii Complexed With Amp
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-12-04 Classification: LIGASE Ligands: SO4, CL, AMP |
 |
Crystal Structure Of Faicar Synthetase (Purp) From M. Jannaschii Complexed With Adp And Faicar
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-12-04 Classification: LIGASE Ligands: SO4, CL, ADP, FAI |
 |
Crystal Structure Of Purp From Pyrococcus Furiosus Complexed With Amp And Aicar
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-12-04 Classification: UNKNOWN FUNCTION |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-12-04 Classification: UNKNOWN FUNCTION |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-12-04 Classification: UNKNOWN FUNCTION Ligands: PO4, NA, ATP, MPD |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-12-04 Classification: UNKNOWN FUNCTION Ligands: PO4, ADP |
 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-10-30 Classification: LYASE Ligands: F2P |
 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-10-30 Classification: LYASE Ligands: 13P |
 |
M. Jannaschii Adh Synthase Complexed With Dihydroxyacetone Phosphate And Glycerol
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-10-30 Classification: LYASE Ligands: 13P, GOL |
 |
Crystal Structure Of An Amide Bond Forming F420-Gamma Glutamyl Ligase From Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2007-05-15 Classification: LIGASE Ligands: MN, ACT, GDP, EDO, GOL |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2007-04-24 Classification: HYDROLASE Ligands: IMP |
 |
Crystal Structure Of Puro/Aicar From Methanothermobacter Thermoautotrophicus
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-04-24 Classification: HYDROLASE Ligands: AMZ |
 |
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-04-24 Classification: HYDROLASE |
 |
Crystal Structure Of Homolog Of F420-0:Gamma-Glutamyl Ligase From Archaeoglobus Fulgidus Reveals A Novel Fold.
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-04-04 Classification: LIGASE |
 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2003-12-09 Classification: LYASE Ligands: SO4 |

