Search Count: 28
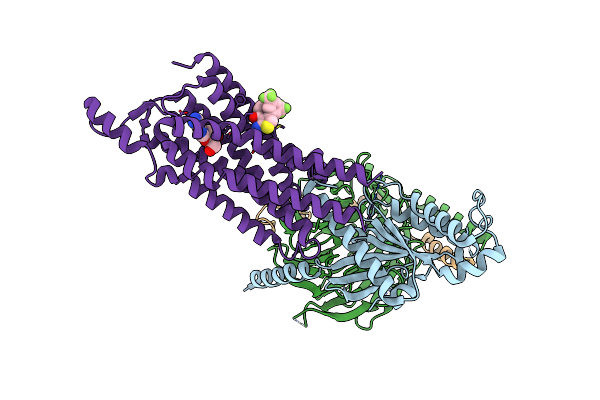 |
Cryo-Em Structure Of The Human Adenosine A1 Receptor-Gi2-Protein Complex Bound To Its Endogenous Agonist And An Allosteric Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: ADN, XTD |
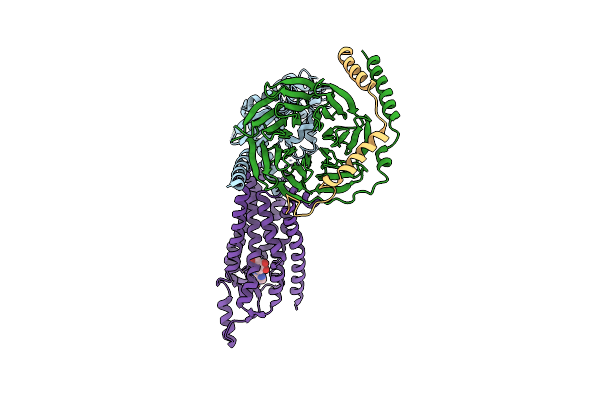 |
Cryo-Em Structure Of The Human Adenosine A1 Receptor-Gi2-Protein Complex Bound To Its Endogenous Agonist
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: SIGNALING PROTEIN Ligands: ADN |
 |
Crystal Structure Of Fab1, The Fab Fragment Of The Anti-Bama Monoclonal Antibody Mab1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2021-06-02 Classification: IMMUNE SYSTEM |
 |
Lateral-Closed Conformation Of The Lid-Locked Bam Complex (Bama E435C S665C, Bambdce) By Cryoem
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: MEMBRANE PROTEIN |
 |
Lateral-Open Conformation Of The Lid-Locked Bam Complex (Bama E435C S665C, Bambdce) By Cryoem
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: MEMBRANE PROTEIN |
 |
Lateral-Open Conformation Of The Lid-Locked Bam Complex (Bama E435C S665C, Bambdce) Bound By A Bactericidal Fab Fragment
Organism: Escherichia coli k-12 , Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: MEMBRANE PROTEIN |
 |
Lateral-Open Conformation Of The Wild-Type Bam Complex (Bamabcde) Bound To A Bactericidal Fab Fragment
Organism: Escherichia coli k-12, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: MEMBRANE PROTEIN |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2021-02-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-02-17 Classification: DE NOVO PROTEIN |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-11-08 Classification: PROTEIN TRANSPORT Ligands: SO4 |
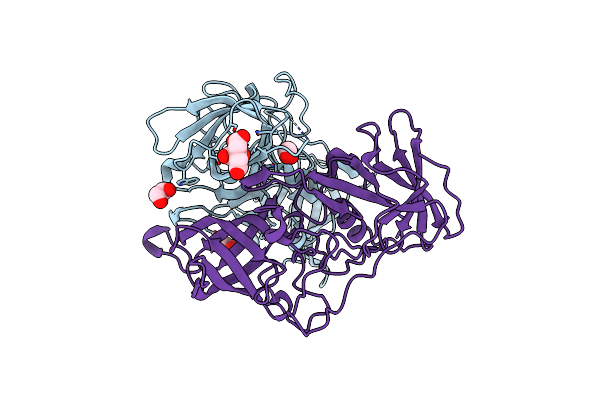 |
Crystal Structure Of Gii.10 P Domain In Complex With Disinfectant Puregreen24
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2015-09-09 Classification: VIRAL PROTEIN Ligands: FLC, EDO |
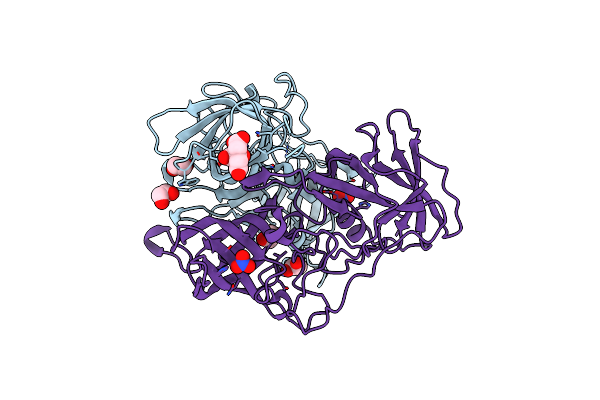 |
Organism: Norwalk virus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-09-09 Classification: VIRAL PROTEIN Ligands: EDO, FLC, NO3 |
 |
Crystal Structure Of Hepatitis C Virus Ns3 Helicase Inhibitor Co-Complex With Fragment 1 [(5-Bromo-1H-Indol-3-Yl)Acetic Acid]
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-03-05 Classification: Hydrolase/Hydrolase Inhibitor Ligands: 2SX, CA |
 |
Crystal Structure Of Hepatitis C Virus Ns3 Helicase Inhibitor Co-Complex With Compound 7 [[1-(3-Chlorobenzyl)-1H-Indol-3-Yl]Acetic Acid]
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-03-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2SY, CA |
 |
Crystal Structure Of Hepatitis C Virus Ns3 Helicase Inhibitor Co-Complex With Compound 9 [1-(3-Ethynylbenzyl)-1H-Indol-3-Yl]Acetic Acid]
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-03-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, 2T2 |
 |
Crystal Structure Of Hepatitis C Virus Ns3 Helicase Inhibitor Co-Complex With Compound 13 [[1-(2-Methoxy-5-Nitrobenzyl)-1H-Indol-3-Yl]Acetic Acid]
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-03-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, 2T7 |
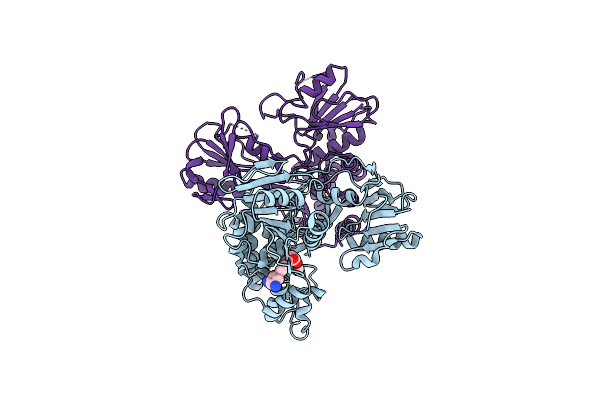 |
Crystal Structure Of Hepatitis C Virus Ns3 Helicase Inhibitor Co-Complex With Compound 19 [[6-(3,5-Diaminophenyl)-1-(2-Methoxy-5-Nitrobenzyl)-1H-Indol-3-Yl]Acetic Acid]
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-03-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2T9, CA |
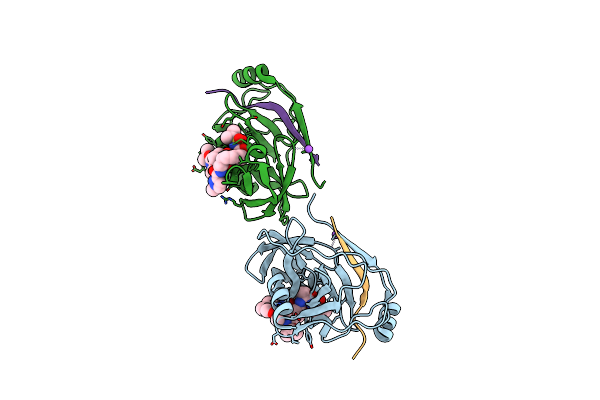 |
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2013-01-02 Classification: Hydrolase/Hydrolase inhibitor |
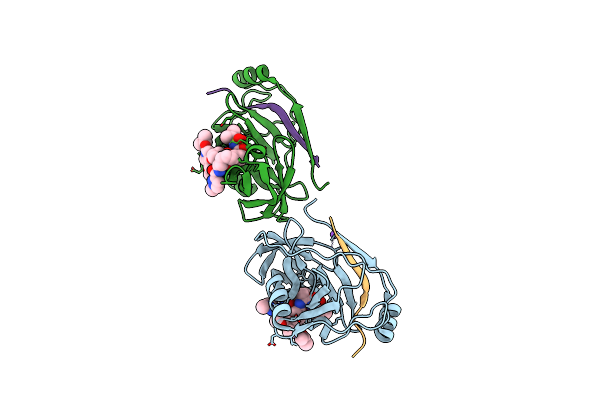 |
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-01-02 Classification: HYDROLASE/HYDROLASE INHIBITOR |
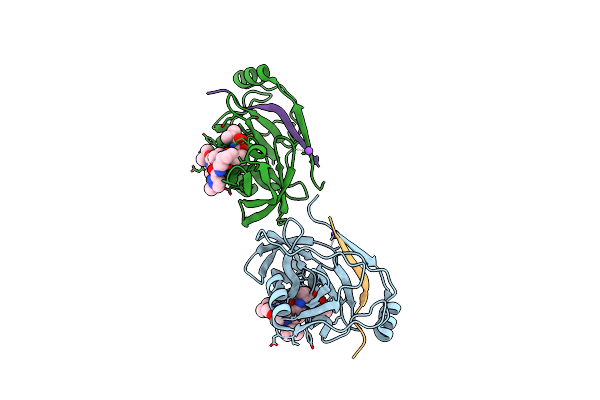 |
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-01-02 Classification: HYDROLASE/HYDROLASE INHIBITOR |

