Search Count: 92
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-03-12 Classification: PROTEIN TRANSPORT Ligands: CL |
 |
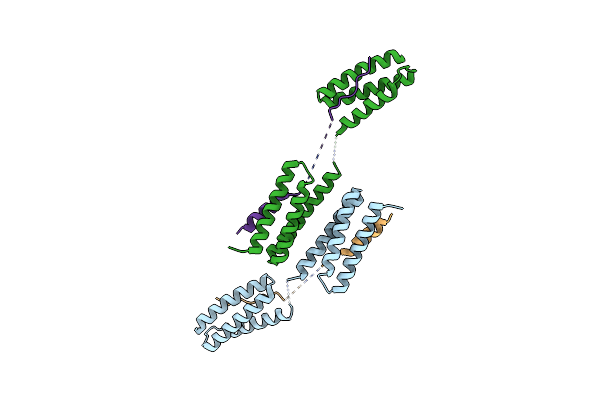
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-31 Classification: SIGNALING PROTEIN Ligands: CA |
 |
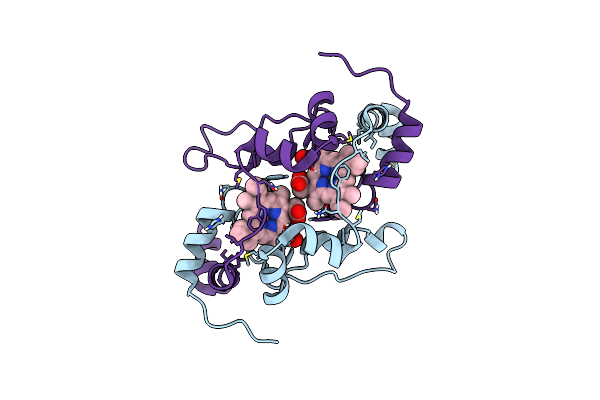
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-10-25 Classification: PEPTIDE BINDING PROTEIN |
 |
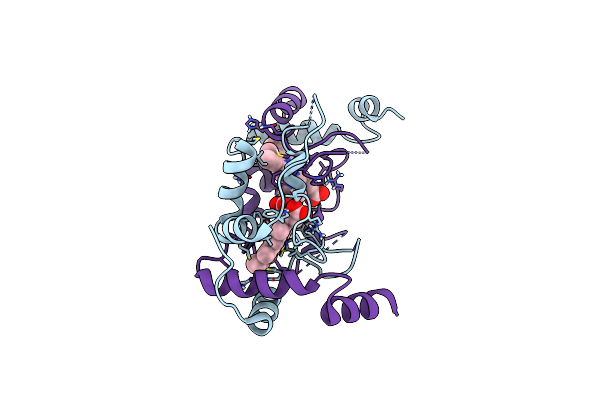
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-05-10 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
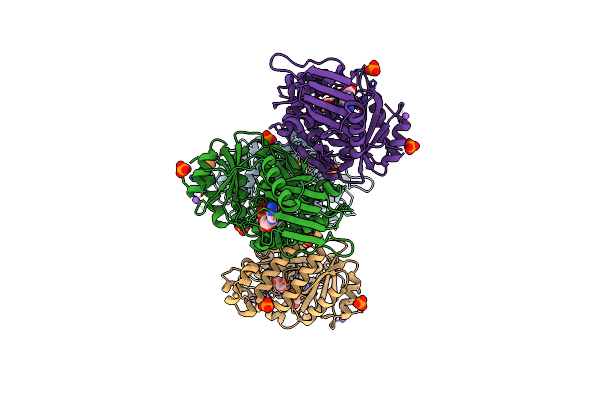
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2023-05-10 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Crystal Structure Of Consomatin-Ro1 <Structure_Details=Consomatin-Ro1, A Cone Snail Venom Sstl Mimetic
Organism: Conus rolani
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-04-13 Classification: TOXIN Ligands: V1J |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-09-29 Classification: SUGAR BINDING PROTEIN Ligands: GLA, ADP, PO4, MG, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-09-29 Classification: SUGAR BINDING PROTEIN Ligands: ADP, GLA, PO4, MG, NA, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-09-29 Classification: Transferase/Inhibitor Ligands: GLA, 4QI, PO4, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-09-29 Classification: Transferase/Inhibitor Ligands: GLA, V3V, NA, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2021-07-21 Classification: PROTEIN FIBRIL |
 |
Crystal Structure Of N-Phenylalanine Peptoid-Modified Collagen Triple Helix
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-07-21 Classification: PROTEIN FIBRIL |
 |
Pie12 D-Peptide Against Hiv Entry (In Complex With Iqn17 Q577R Resistance Mutant)
Organism: Synthetic construct, Saccharomyces cerevisiae, Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-02-05 Classification: VIRAL PROTEIN/INHIBITOR Ligands: CL |
 |
Crystal Structure Of Zbtb38 C-Terminal Zinc Fingers 6-9 In Complex With Methylated Dna
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-11-07 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Crystal Structure Of Zbtb38 C-Terminal Zinc Fingers 6-9 K1055R In Complex With Methylated Dna
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-11-07 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-10-25 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-09-20 Classification: TRANSCRIPTION Ligands: IPA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-09 Classification: DE NOVO PROTEIN Ligands: GOL |
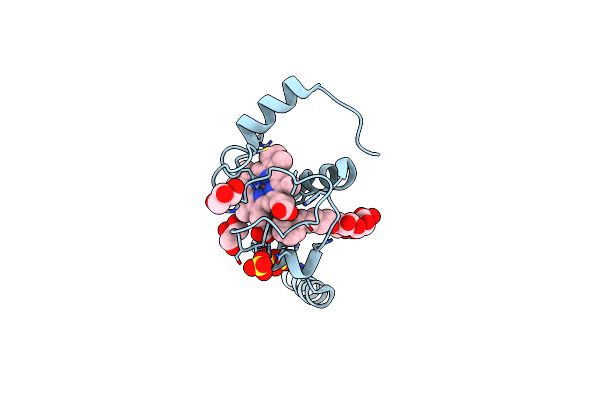 |
Crystal Structure Of A Domain-Swapped Dimer Of Yeast Iso-1-Cytochrome C With Cymal5
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-22 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4, CM5, CL, GOL |
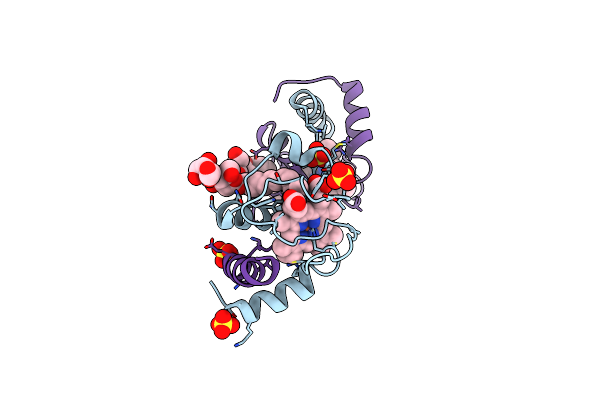 |
Crystal Structure Of A Domain-Swapped Dimer Of Yeast Iso-1-Cytochrome C With Omega-Undecylenyl-Beta-D-Maltopyranoside
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-03-22 Classification: ELECTRON TRANSPORT Ligands: HEC, 6UZ, SO4 |

