Search Count: 15
 |
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex In The Ca2+ Bound State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex In The Ca2+ Free State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: K, CA |
 |
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex Bound To The Bee Toxin Apamin
Organism: Homo sapiens, Apis mellifera
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN/TOXIN Ligands: K, CA |
 |
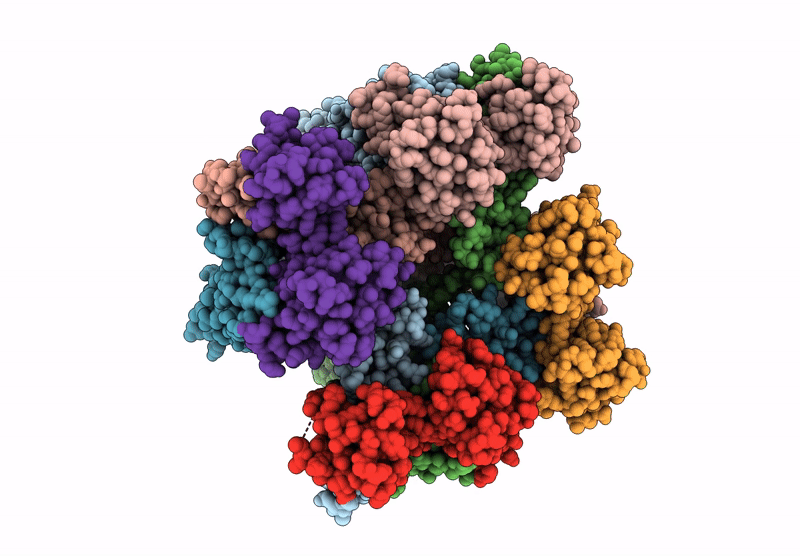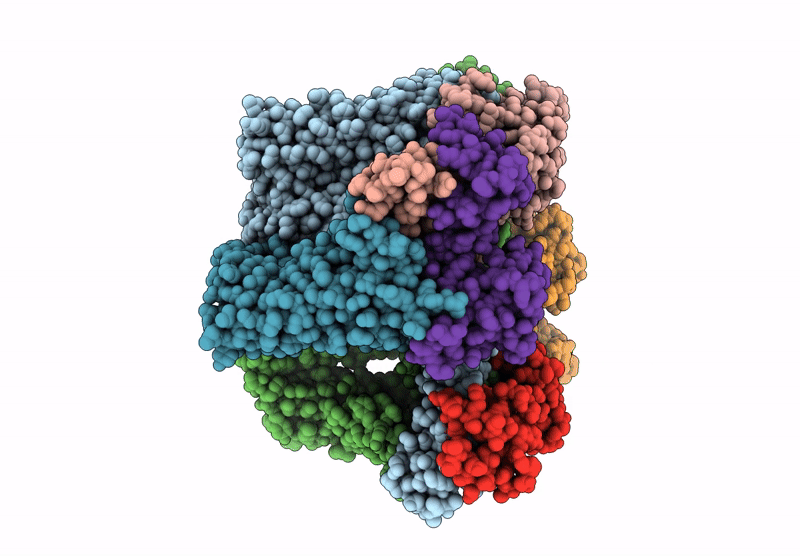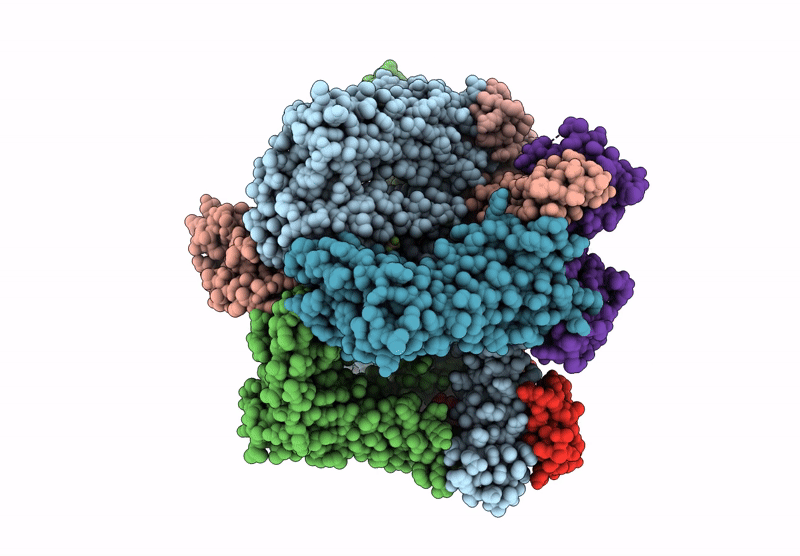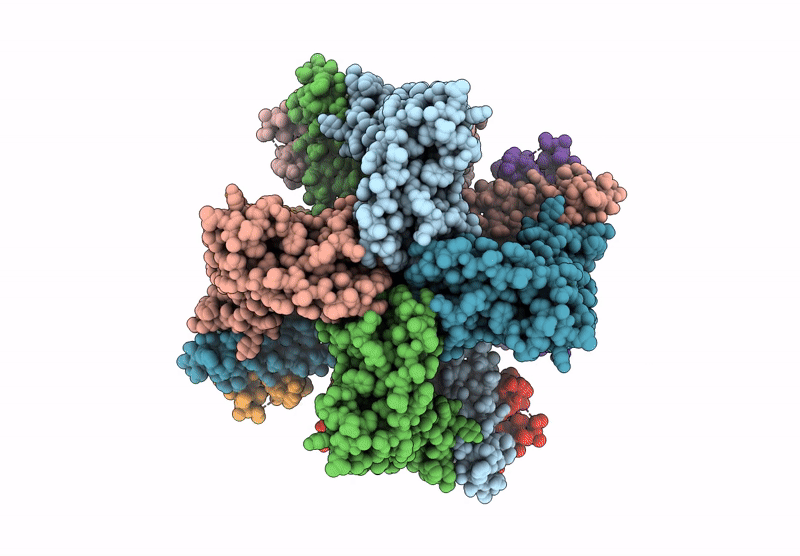
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex Bound To A Small Molecule Inhibitor
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: A1B8D, K, CA |
 |
Cryo-Em Structure Of The Human Sk2-4 Chimera/Calmodulin Channel Complex Bound To A Small Molecule Activator
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: A1B8G, K, CA |
 |
Structure Of The Voltage-Gated Sodium Channel Navpas From American Cockroach Periplaneta Americana In Complex With Scorpion Alpha-Toxin Lqhait
Organism: Periplaneta americana, Leiurus quinquestriatus hebraeus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Single Particle Cryo-Em Structure Of The Voltage-Gated K+ Channel Eag1 3-13 Deletion Mutant Bound To Calmodulin (Conformation 2)
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: TRANSPORT PROTEIN/CALCIUM BINDING PROTEIN |
 |
Single Particle Cryo-Em Structure Of The Voltage-Gated K+ Channel Eag1 3-13 Deletion Mutant Bound To Calmodulin (Conformation 1)
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: TRANSPORT PROTEIN/CALCIUM BINDING PROTEIN |
 |
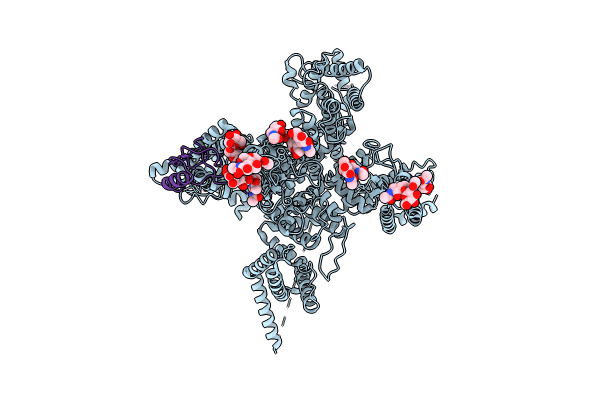
Single Particle Cryo-Em Structure Of The Voltage-Gated K+ Channel Eag1 Bound To The Channel Inhibitor Calmodulin
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.78 Å Release Date: 2016-08-17 Classification: METAL TRANSPORT/CALCIUM BINDING PROTEIN Ligands: NAG, Y01 |
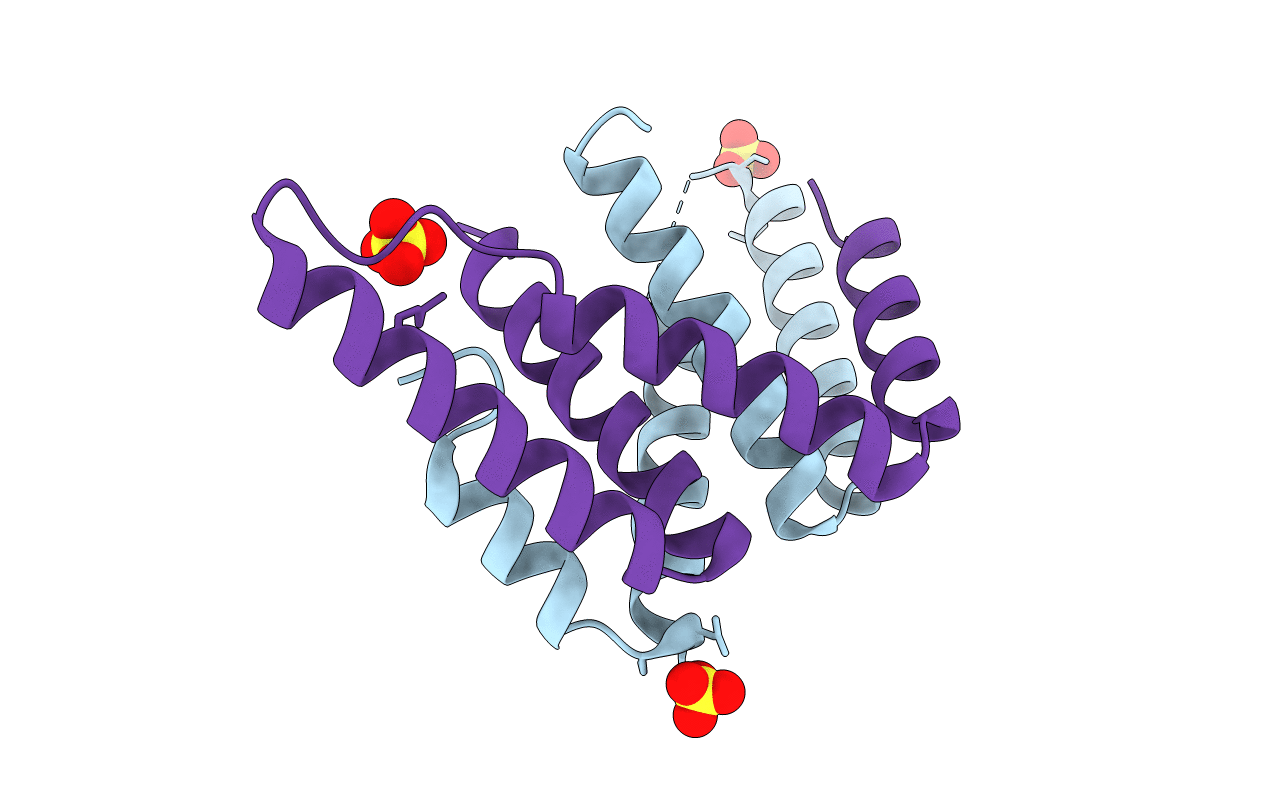 |
Structure Of A Class 2 Docking Domain Complex From Modules Curg And Curh Of The Curacin A Polyketide Synthase
Organism: Moorea producens 3l
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-01-29 Classification: PROTEIN BINDING Ligands: SO4 |
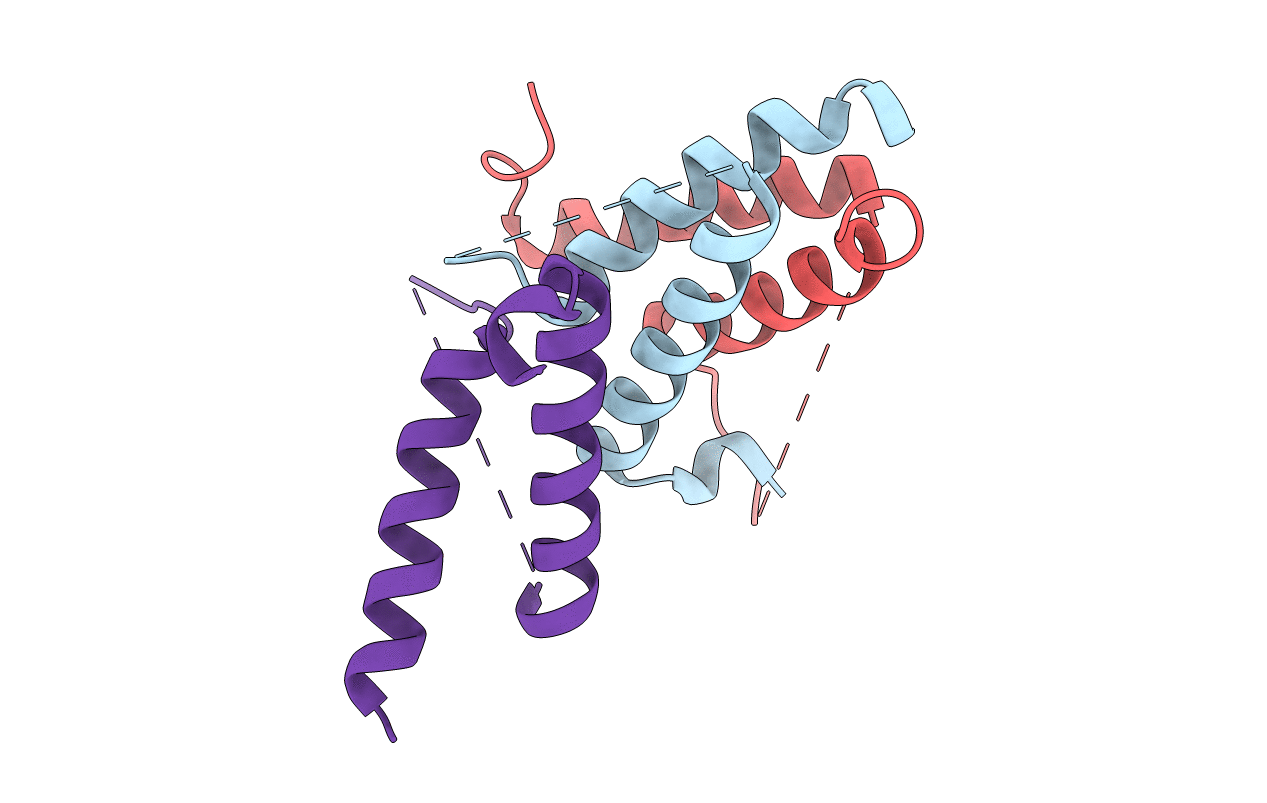 |
Structure Of A Class 2 Docking Domain Complex From Modules Curk And Curl Of The Curacin A Polyketide Synthase
Organism: Moorea producens 3l
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-01-29 Classification: PROTEIN BINDING |
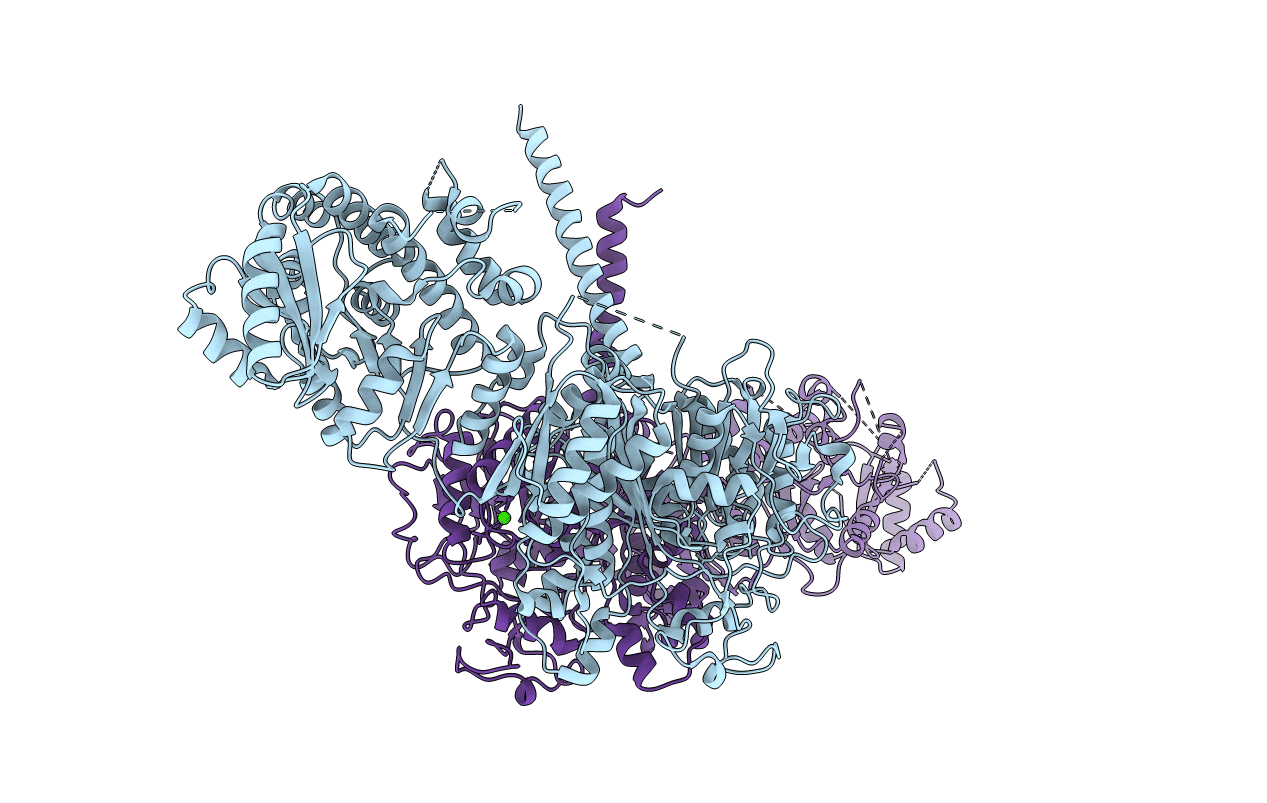 |
Structure Of A Ketosynthase-Acyltransferase Di-Domain From Module Curl Of The Curacin A Polyketide Synthase
Organism: Moorea producens 3l
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-01-29 Classification: TRANSFERASE Ligands: CA |
 |
Plmkr1-Ketoreductase From The First Module Of Phoslactomycin Biosynthesis In Streptomyces Sp. Hk803
Organism: Streptomyces sp. hk803
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2013-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, ACA |
 |
Redj-Thioesterase From The Prodiginine Biosynthetic Pathway In Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2011-05-04 Classification: HYDROLASE |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-05-04 Classification: HYDROLASE Ligands: PG4 |

