Search Count: 28
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: SO4, GOL, ACT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: CL, GOL, U5P |
 |
Solution Structural Studies Of Gtp:Adenosylcobinamide-Phosphate Guanylyltransferase (Coby) From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii
Method: SOLUTION NMR Release Date: 2015-11-11 Classification: TRANSFERASE |
 |
X-Ray Structure Of Ester Chemical Analogue [O-Ile50,O-Ile50']Hiv-1 Protease Complexed With Mvt-101
Method: X-RAY DIFFRACTION
Resolution:1.50 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: 2NC |
 |
X-Ray Structure Of Ester Chemical Analogue [O-Ile50,O-Ile50']Hiv-1 Protease Complexed With Kvs-1 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.90 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: SO4, KVS |
 |
X-Ray Structure Of Ester Chemical Analogue 'Covalent Dimer' [Ile50,O-Ile50']Hiv-1 Protease Complexed With Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.61 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: 2NC, SO4 |
 |
X-Ray Structure Of Ester Chemical Analogue 'Covalent Dimer' [Ile50,O-Ile50']Hiv-1 Protease Complexed With Kvs-1 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.80 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: KVS, SO4 |
 |
X-Ray Structure Of Ester Chemical Analogue [O-Gly51,O-Gly51']Hiv-1 Protease Complexed With Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.45 Å Release Date: 2011-11-02 Classification: hydrolase/hydrolase inhibitor Ligands: 2NC, SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2011-10-12 Classification: CHAPERONE |
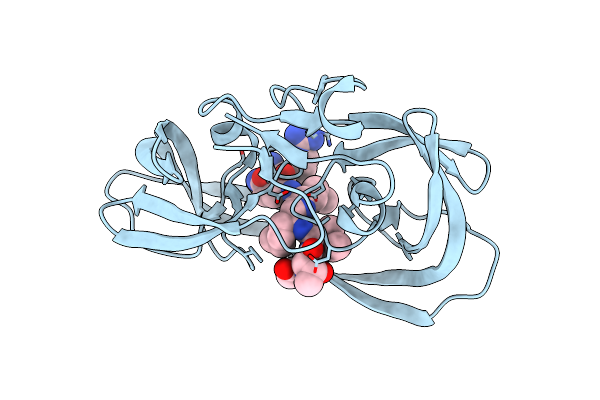 |
Crystal Structure Of Chemically Synthesized 'Covalent Dimer' [Gly51/D-Ala51']Hiv-1 Protease
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2011-07-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2011-06-15 Classification: CHAPERONE |
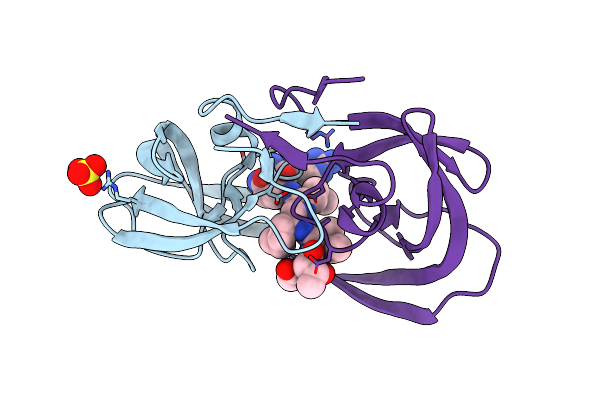 |
Crystal Structure Of Chemically Synthesized Hiv-1 Protease With Reduced Isostere Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.30 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
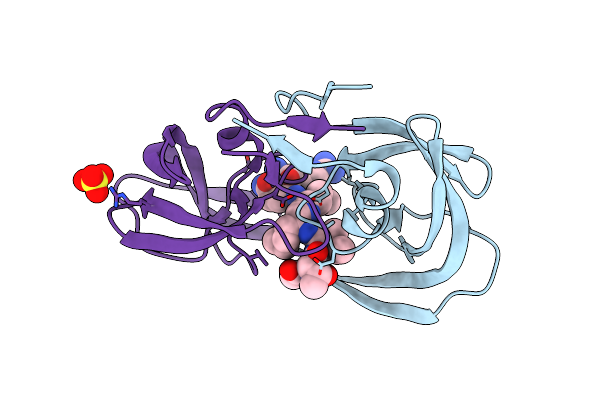 |
Crystal Structure Of [L-Ala51/51']Hiv-1 Protease With Reduced Isostere Mvt-101 Inhibitor
Method: X-RAY DIFFRACTION
Resolution:1.30 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
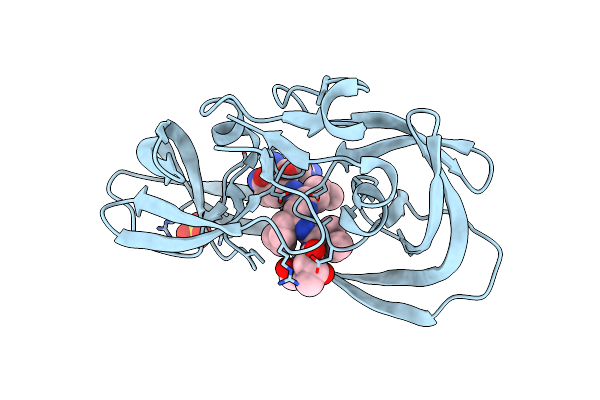 |
Crystal Structure Of A Chemically Synthesized 203 Amino Acid 'Covalent Dimer' [Gly51;Aib51']Hiv-1 Protease Molecule Complexed With Mvt-101 Reduced Isostere Inhibitor At 1.6 A Resolution
Method: X-RAY DIFFRACTION
Resolution:1.61 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC, SO4 |
 |
Crystal Structure Of Chemically Synthesized 203 Amino Acid 'Covalent Dimer' [L-Ala;Gly51']Hiv-1 Protease Molecule Complexed With Mvt-101 Reduced Isostere Inhibitor At 1.4 A Resolution
Method: X-RAY DIFFRACTION
Resolution:1.40 Å Release Date: 2011-04-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |
 |
Method: X-RAY DIFFRACTION
Resolution:1.71 Å Release Date: 2010-05-26 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |
 |
Method: X-RAY DIFFRACTION
Resolution:1.80 Å Release Date: 2010-04-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |
 |
Crystal Structure Of A Chemically Synthesized 203 Amino Acid 'Covalent Dimer' [L-Ala51,D-Ala51'] Hiv-1 Protease Molecule
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2010-01-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 2NC |
 |
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2009-11-10 Classification: SIGNALING PROTEIN |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-09-22 Classification: ISOMERASE Ligands: FAD, GDU |

