Search Count: 71
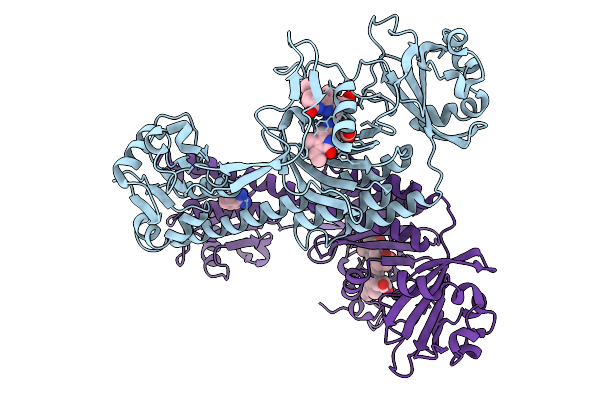 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 3 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
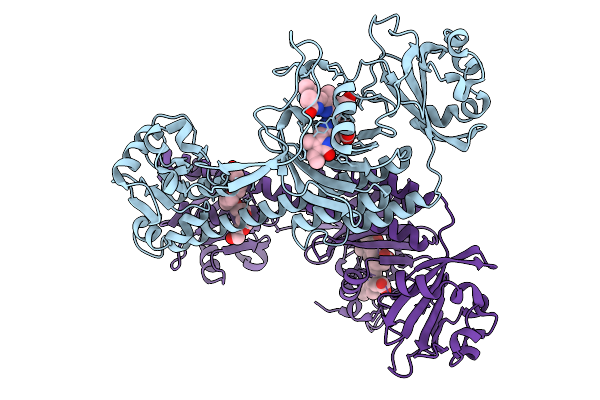 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
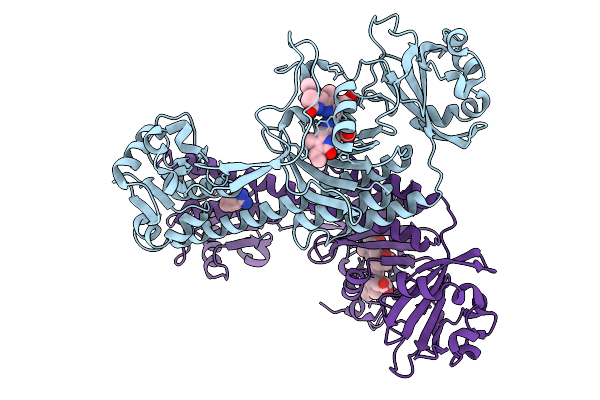 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
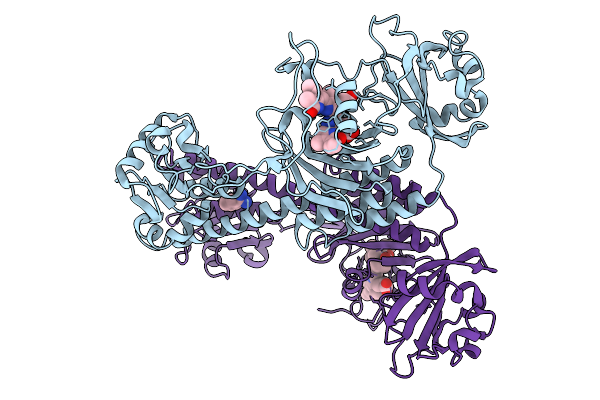 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (-68 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (-40 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (-33 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (-2 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (34 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (72 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (109 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (144 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (166 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (207 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (233 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (280 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (295 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (344 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |
 |
Crystal Structure Of Pas-Gaf Domain Of The Phytochrome From Deinococcus Radiodurans At Femto And Picosecond Timescales (363 Fs)
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: LBV |

