Search Count: 38
 |
Structure Of An Hxlr/Duf24 Family Transcription Regulator, Cdtr_3200 From Hypervirulent Clostridioides Difficile R20291
Organism: Clostridioides difficile (strain r20291)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-09-15 Classification: DNA BINDING PROTEIN |
 |
Role Of The Highly Conserved G68 Residue In The Yeast Phosphorelay Protein Ypd1: Implications For Interactions Between Histidine Phosphotransfer (Hpt) And Response Regulator Proteins
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-02-06 Classification: TRANSFERASE |
 |
Co-Crystal Structure Of The Saccharomyces Cerevisiae Histidine Phosphotransfer Signaling Protein Ypd1 And The Receiver Domain Of Its Downstream Response Regulator Ssk1
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-06-07 Classification: SIGNALING PROTEIN Ligands: GOL, PO4 |
 |
Crystal Structure Of A Padr Family Transcription Regulator From Hypervirulent Clostridium Difficile R20291 - Cdpadr_0991 To 1.89 Angstrom Resolution
Organism: Peptoclostridium difficile (strain r20291)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2016-10-05 Classification: DNA BINDING PROTEIN |
 |
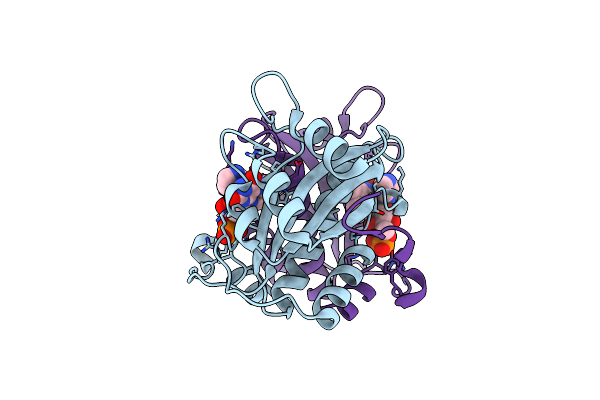
Fmn-Dependent Nitroreductase (Cdr20291_0684) From Clostridium Difficile R20291
Organism: Peptoclostridium difficile (strain r20291)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, PO4 |
 |
Fmn-Dependent Nitroreductase (Cdr20291_0767) From Clostridium Difficile R20291
Organism: Peptoclostridium difficile (strain r20291)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-09-21 Classification: OXIDOREDUCTASE Ligands: FMN, IMD |
 |
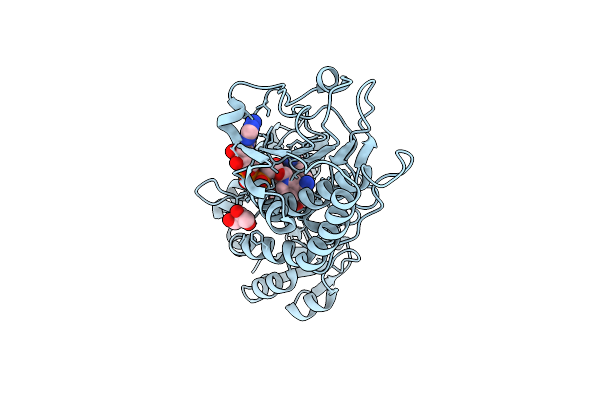
Crystal Structure Of C205S Mutant And Saccharopine Dehydrogenase From Saccharomyces Cerevisiae.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2012-02-01 Classification: OXIDOREDUCTASE |
 |
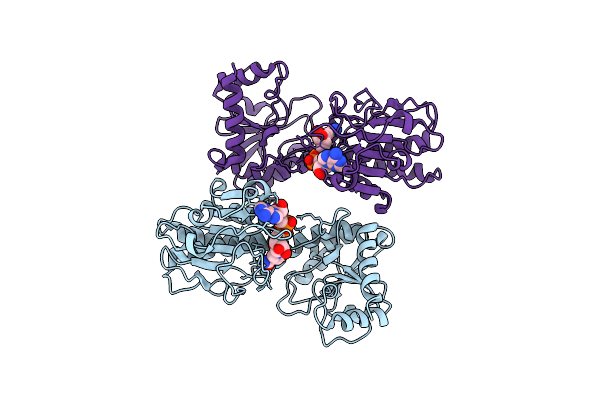
Crystal Structure Of Saccharopine Dehydrogenase From Saccharomyces Cerevisiae With Bound Saccharopine And Nadh
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2012-02-01 Classification: OXIDOREDUCTASE Ligands: NAI, SHR, GOL |
 |
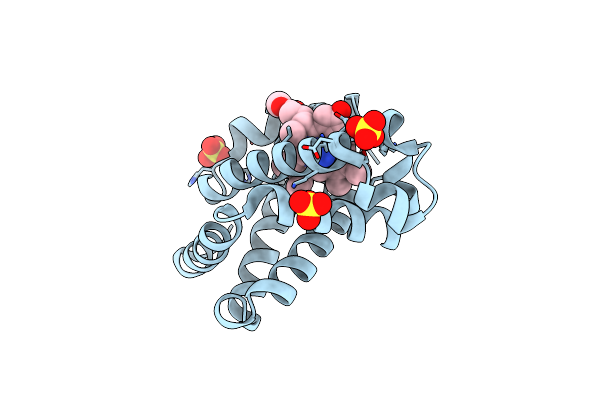
Crystal Structure Of Saccharopine Dehydrogenase From Saccharomyces Cervisiae Complexed With Nad.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-02-01 Classification: OXIDOREDUCTASE Ligands: NAD, CL |
 |
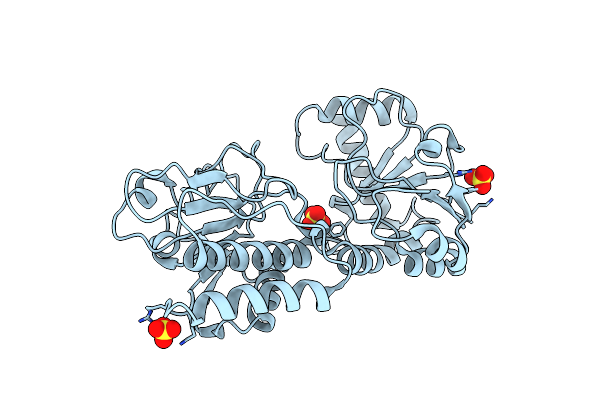
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-11-11 Classification: TRANSPORT PROTEIN Ligands: SO4, CYN, HKL |
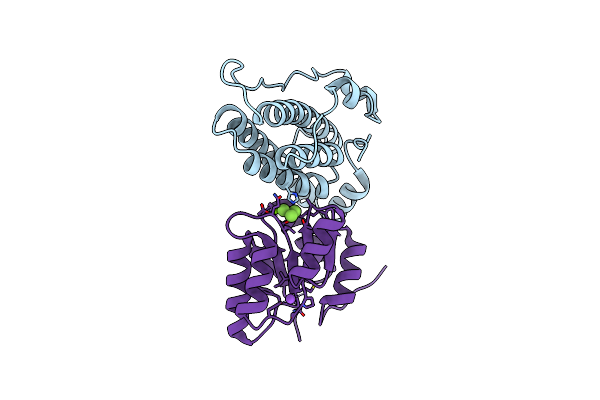 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-01-15 Classification: Signaling PROTEIN/Transferase Ligands: MG, NA, BEF |
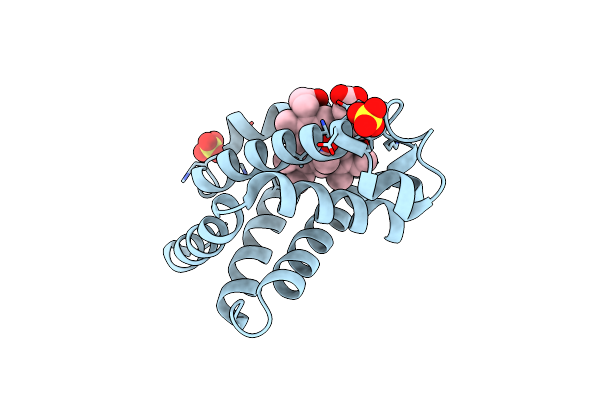 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-11-06 Classification: OXYGEN STORAGE/TRANSPORT Ligands: SO4, HEM, NSM |
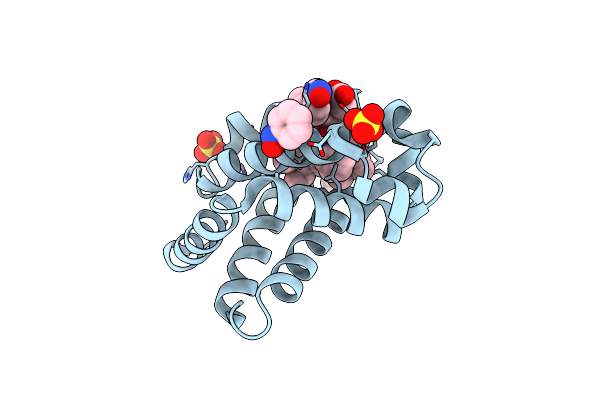 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-11-06 Classification: OXYGEN STORAGE/TRANSPORT Ligands: SO4, HEM, NBE |
 |
Crystal Structure Of Sulfate-Bound Saccharopine Dehydrogenase (L-Lys Forming) From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-10-30 Classification: OXIDOREDUCTASE Ligands: SO4, CL |
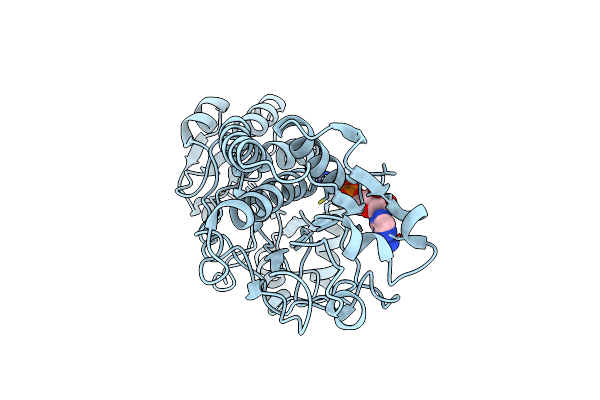 |
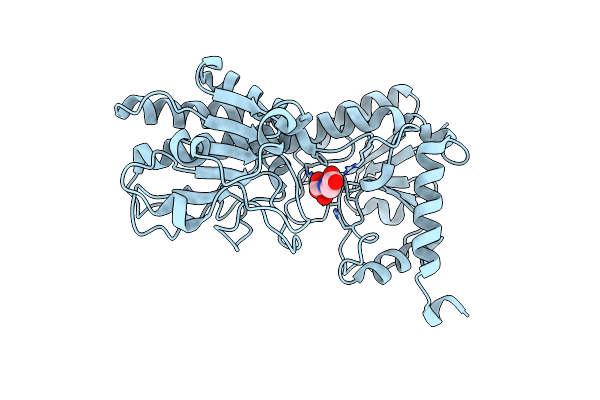
Crystal Structure Of Amp-Bound Saccharopine Dehydrogenase (L-Lys Forming) From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-10-30 Classification: OXIDOREDUCTASE Ligands: AMP |
 |
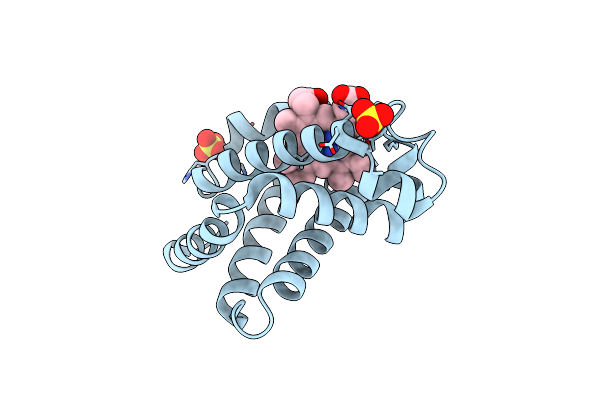
Crystal Structure Of Oxalylglycine-Bound Saccharopine Dehydrogenase (L-Lys Forming) From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-10-30 Classification: OXIDOREDUCTASE Ligands: OGA |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2007-10-16 Classification: OXYGEN STORAGE/TRANSPORT Ligands: SO4, MNR |
 |
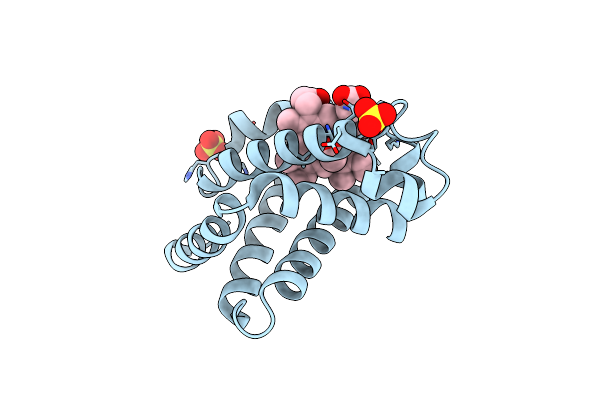
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-16 Classification: OXYGEN STORAGE/TRANSPORT Ligands: SO4, MNH |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2007-10-16 Classification: OXYGEN STORAGE/TRANSPORT Ligands: SO4, MNR, MOH |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2007-10-16 Classification: OXYGEN STORAGE/TRANSPORT Ligands: AZI, SO4, MNR |

