Search Count: 35
 |
Organism: Legionella pneumophila str. lens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TOXIN |
 |
Organism: Legionella pneumophila
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TOXIN |
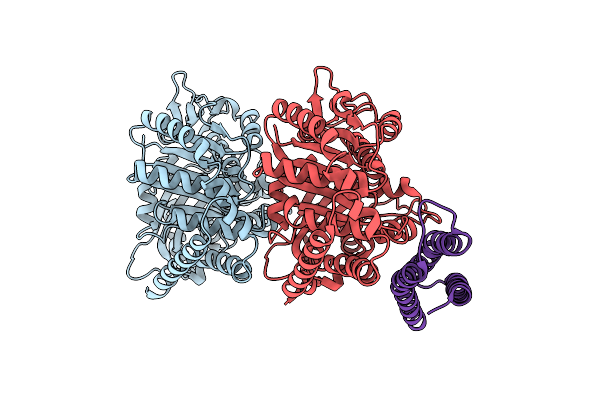 |
Organism: Bos taurus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: CELL CYCLE |
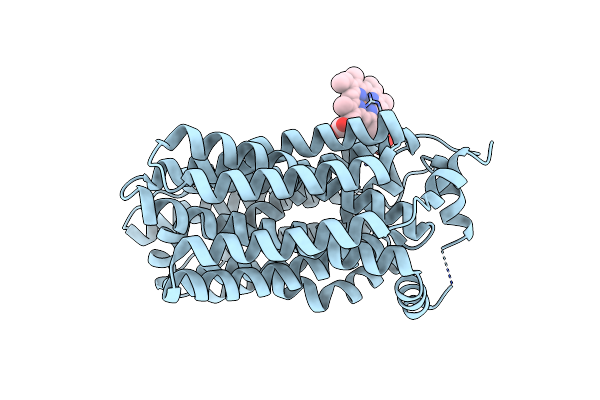 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN Ligands: HEM |
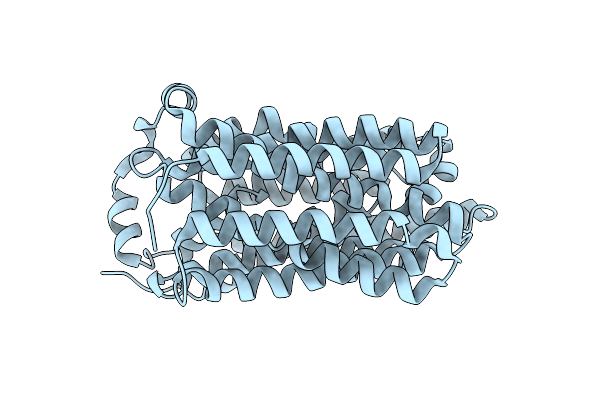 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN |
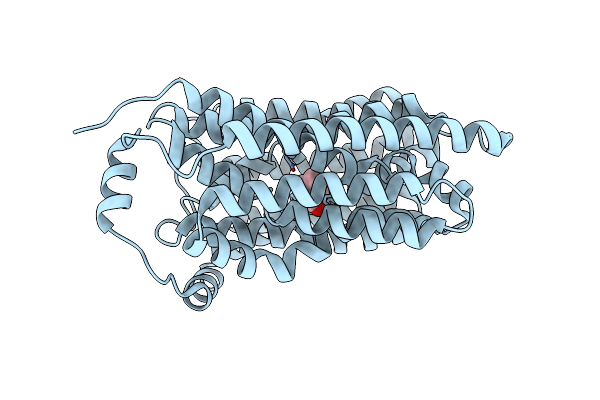 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CHT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CHT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: ETA |
 |
Organism: Epilithonimonas lactis
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN |
 |
Bacterial Sting From Riemerella Anatipestifer In Complex With 3'3'-C-Di-Gmp
Organism: Riemerella anatipestifer yb2
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: C2E, CA |
 |
Organism: Riemerella anatipestifer yb2
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-01-10 Classification: SIGNALING PROTEIN Ligands: PG4 |
 |
Organism: Larkinella arboricola
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2024-01-03 Classification: SIGNALING PROTEIN Ligands: C2E |
 |
Organism: Epilithonimonas lactis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-01-03 Classification: SIGNALING PROTEIN Ligands: 2BA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-01-27 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Complex Of Sars-Cov-2 Spike Protein And Fab P17 With One Rbd In Open State And Two Rbd In Closed State
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-16 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-16 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-16 Classification: VIRAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2020-12-02 Classification: PROTEIN TRANSPORT Ligands: GOL, SO4, MES |

